Overexpression of a Novel Cytochrome P450 Promotes Flavonoid Biosynthesis and Osmotic Stress Tolerance in Transgenic Arabidopsis
- PMID: 31561549
- PMCID: PMC6826380
- DOI: 10.3390/genes10100756
Overexpression of a Novel Cytochrome P450 Promotes Flavonoid Biosynthesis and Osmotic Stress Tolerance in Transgenic Arabidopsis
Abstract
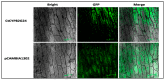
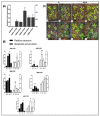
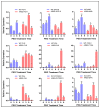
Flavonoids are mainly associated with growth, development, and responses to diverse abiotic stresses in plants. A growing amount of data have demonstrated the biosynthesis of flavonoids through multienzyme complexes of which the membrane-bounded cytochrome P450 supergene family shares a crucial part. However, the explicit regulation mechanism of Cytochrome P450s related to flavonoid biosynthesis largely remains elusive. In the present study, we reported the identification of a stress-tolerant flavonoid biosynthetic CtCYP82G24 gene from Carthamus tinctorius. The transient transformation of CtCYP82G24 determined the subcellular localization to the cytosol. Heterologously expressed CtCYP82G24 was effective to catalyze the substrate-specific conversion, promoting the de novo biosynthesis of flavonoids in vitro. Furthermore, a qRT-PCR assay and the accumulation of metabolites demonstrated that the expression of CtCYP82G24 was effectively induced by Polyethylene glycol stress in transgenic Arabidopsis. In addition, the overexpression of CtCYP82G24 could also trigger expression levels of several other flavonoid biosynthetic genes in transgenic plants. Taken together, our findings suggest that CtCYP82G24 overexpression plays a decisive regulatory role in PEG-induced osmotic stress tolerance and alleviates flavonoid accumulation in transgenic Arabidopsis.
Keywords: Cytochrome P450; abiotic stress; flavonoid biosynthesis; heterologous expression; transgenic Arabidopsis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Stafford H. Recent Advances in Phytochemistry. Volume 8. Elsevier; Warsaw, Portland: 1974. Possible multienzyme complexes regulating the formation of C6-C3 phenolic compounds and lignins in higher plants; pp. 53–79.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

