Urothelial organoids originating from Cd49fhigh mouse stem cells display Notch-dependent differentiation capacity
- PMID: 31562298
- PMCID: PMC6764959
- DOI: 10.1038/s41467-019-12307-1
Urothelial organoids originating from Cd49fhigh mouse stem cells display Notch-dependent differentiation capacity
Abstract
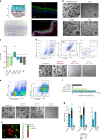
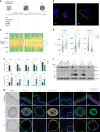
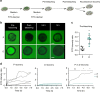
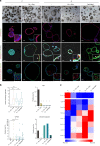
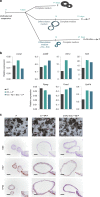
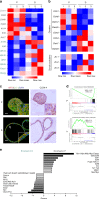
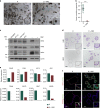
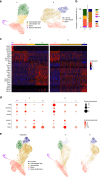
Understanding urothelial stem cell biology and differentiation has been limited by the lack of methods for their unlimited propagation. Here, we establish mouse urothelial organoids that can be maintained uninterruptedly for >1 year. Organoid growth is dependent on EGF and Wnt activators. High CD49f/ITGA6 expression features a subpopulation of organoid-forming cells expressing basal markers. Upon differentiation, multilayered organoids undergo reduced proliferation, decreased cell layer number, urothelial program activation, and acquisition of barrier function. Pharmacological modulation of PPARγ and EGFR promotes differentiation. RNA sequencing highlighted genesets enriched in proliferative organoids (i.e. ribosome) and transcriptional networks involved in differentiation, including expression of Wnt ligands and Notch components. Single-cell RNA sequencing (scRNA-Seq) analysis of the organoids revealed five clusters with distinct gene expression profiles. Together, with the use of γ-secretase inhibitors and scRNA-Seq, confirms that Notch signaling is required for differentiation. Urothelial organoids provide a powerful tool to study cell regeneration and differentiation.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Wu X-R, Manabe M, Jun Y, Sun T-T. Large scale purification and immunolocalization of bovine uroplakins I, II, and III. J. Biol. Chem. 1990;265:19170–19179. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

