FIERY1 promotes microRNA accumulation by suppressing rRNA-derived small interfering RNAs in Arabidopsis
- PMID: 31562313
- PMCID: PMC6765019
- DOI: 10.1038/s41467-019-12379-z
FIERY1 promotes microRNA accumulation by suppressing rRNA-derived small interfering RNAs in Arabidopsis
Abstract
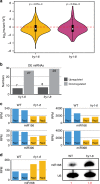
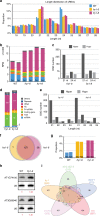
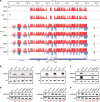
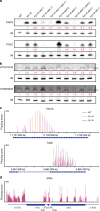
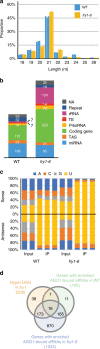
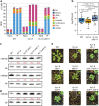
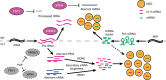
Plant microRNAs (miRNAs) associate with ARGONAUTE1 (AGO1) to direct post-transcriptional gene silencing and regulate numerous biological processes. Although AGO1 predominantly binds miRNAs in vivo, it also associates with endogenous small interfering RNAs (siRNAs). It is unclear whether the miRNA/siRNA balance affects miRNA activities. Here we report that FIERY1 (FRY1), which is involved in 5'-3' RNA degradation, regulates miRNA abundance and function by suppressing the biogenesis of ribosomal RNA-derived siRNAs (risiRNAs). In mutants of FRY1 and the nuclear 5'-3' exonuclease genes XRN2 and XRN3, we find that a large number of 21-nt risiRNAs are generated through an endogenous siRNA biogenesis pathway. The production of risiRNAs correlates with pre-rRNA processing defects in these mutants. We also show that these risiRNAs are loaded into AGO1, causing reduced loading of miRNAs. This study reveals a previously unknown link between rRNA processing and miRNA accumulation.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Arabidopsis FIERY1, XRN2, and XRN3 are endogenous RNA silencing suppressors.Plant Cell. 2007 Nov;19(11):3451-61. doi: 10.1105/tpc.107.055319. Epub 2007 Nov 9. Plant Cell. 2007. PMID: 17993620 Free PMC article.
-
Activity and roles of Arabidopsis thaliana XRN family exoribonucleases in noncoding RNA pathways.J Plant Res. 2017 Jan;130(1):25-31. doi: 10.1007/s10265-016-0887-z. Epub 2016 Dec 17. J Plant Res. 2017. PMID: 27988817 Review.
-
The Slicer Activity of ARGONAUTE1 Is Required Specifically for the Phasing, Not Production, of Trans-Acting Short Interfering RNAs in Arabidopsis.Plant Cell. 2016 Jul;28(7):1563-80. doi: 10.1105/tpc.16.00121. Epub 2016 Jun 27. Plant Cell. 2016. PMID: 27354557 Free PMC article.
-
The miRNA pathway limits AGO1 availability during siRNA-mediated PTGS defense against exogenous RNA.Nucleic Acids Res. 2011 Nov;39(21):9339-44. doi: 10.1093/nar/gkr590. Epub 2011 Aug 3. Nucleic Acids Res. 2011. PMID: 21813456 Free PMC article.
-
A new layer of rRNA regulation by small interference RNAs and the nuclear RNAi pathway.RNA Biol. 2017 Nov 2;14(11):1492-1498. doi: 10.1080/15476286.2017.1341034. Epub 2017 Jul 21. RNA Biol. 2017. PMID: 28640690 Free PMC article. Review.
Cited by
-
Beyond Loading: Functions of Plant ARGONAUTE Proteins.Int J Mol Sci. 2023 Nov 7;24(22):16054. doi: 10.3390/ijms242216054. Int J Mol Sci. 2023. PMID: 38003244 Free PMC article. Review.
-
Uridylation and the SKI complex orchestrate the Calvin cycle of photosynthesis through RNA surveillance of TKL1 in Arabidopsis.Proc Natl Acad Sci U S A. 2022 Sep 20;119(38):e2205842119. doi: 10.1073/pnas.2205842119. Epub 2022 Sep 12. Proc Natl Acad Sci U S A. 2022. PMID: 36095196 Free PMC article.
-
Intragenic heterochromatin-mediated alternative polyadenylation modulates miRNA and pollen development in rice.New Phytol. 2021 Oct;232(2):835-852. doi: 10.1111/nph.17635. Epub 2021 Aug 8. New Phytol. 2021. PMID: 34289124 Free PMC article.
-
The Use of Arabidopsis thaliana to Characterize the Production and Action Stages of the Plant MicroRNA Pathway.Methods Mol Biol. 2025;2900:1-42. doi: 10.1007/978-1-0716-4398-3_1. Methods Mol Biol. 2025. PMID: 40380051
-
RNA interference and turnover in plants -a complex partnership.Front Plant Sci. 2025 Jul 1;16:1608888. doi: 10.3389/fpls.2025.1608888. eCollection 2025. Front Plant Sci. 2025. PMID: 40666303 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

