Miro clusters regulate ER-mitochondria contact sites and link cristae organization to the mitochondrial transport machinery
- PMID: 31562315
- PMCID: PMC6764964
- DOI: 10.1038/s41467-019-12382-4
Miro clusters regulate ER-mitochondria contact sites and link cristae organization to the mitochondrial transport machinery
Abstract
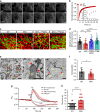
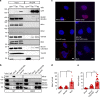
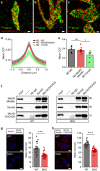
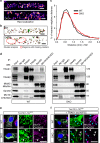
Mitochondrial Rho (Miro) GTPases localize to the outer mitochondrial membrane and are essential machinery for the regulated trafficking of mitochondria to defined subcellular locations. However, their sub-mitochondrial localization and relationship with other critical mitochondrial complexes remains poorly understood. Here, using super-resolution fluorescence microscopy, we report that Miro proteins form nanometer-sized clusters along the mitochondrial outer membrane in association with the Mitochondrial Contact Site and Cristae Organizing System (MICOS). Using knockout mouse embryonic fibroblasts we show that Miro1 and Miro2 are required for normal mitochondrial cristae architecture and Endoplasmic Reticulum-Mitochondria Contacts Sites (ERMCS). Further, we show that Miro couples MICOS to TRAK motor protein adaptors to ensure the concerted transport of the two mitochondrial membranes and the correct distribution of cristae on the mitochondrial membrane. The Miro nanoscale organization, association with MICOS complex and regulation of ERMCS reveal new levels of control of the Miro GTPases on mitochondrial functionality.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

