RBX1 prompts degradation of EXO1 to limit the homologous recombination pathway of DNA double-strand break repair in G1 phase
- PMID: 31562368
- PMCID: PMC7205894
- DOI: 10.1038/s41418-019-0424-4
RBX1 prompts degradation of EXO1 to limit the homologous recombination pathway of DNA double-strand break repair in G1 phase
Abstract
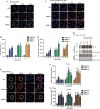
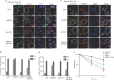
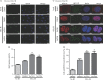
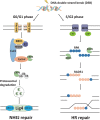
End resection of DNA double-strand breaks (DSBs) to form 3' single-strand DNA (ssDNA) is critical to initiate the homologous recombination (HR) pathway of DSB repair. HR pathway is strictly limited in the G1-phase cells because of lack of homologous DNA as the templates. Exonuclease 1 (EXO1) is the key molecule responsible for 3' ssDNA formation of DSB end resection. We revealed that EXO1 is inactivated in G1-phase cells via ubiquitination-mediated degradation, resulting from an elevated expression level of RING-box protein 1 (RBX1) in G1 phase. The increased RBX1 significantly prompted the neddylation of Cullin1 and contributed to the G1 phase-specific degradation of EXO1. Knockdown of RBX1 remarkedly attenuated the degradation of EXO1 and increased the end resection and HR activity in γ-irradiated G1-phase cells, as demonstrated by the increased formation of RPA32, BrdU, and RAD51 foci. And EXO1 depletion mitigated DNA repair defects due to RBX1 reduction. Moreover, increased autophosphorylation of DNA-PKcs at S2056 was found to be responsible for the higher expression level of the RBX1 in the G1 phase. Inactivation of DNA-PKcs decreased RBX1 expression, and simultaneously increased EXO1 expression and DSB end resection in G1-phase cells. This study demonstrates a new mechanism for restraining the HR pathway of DNA DSB repair in G1 phase via RBX1-prompted inactivation of EXO1.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures





Similar articles
-
DNA end resection is needed for the repair of complex lesions in G1-phase human cells.Cell Cycle. 2014;13(16):2509-16. doi: 10.4161/15384101.2015.941743. Cell Cycle. 2014. PMID: 25486192 Free PMC article.
-
Phosphorylation of Exo1 modulates homologous recombination repair of DNA double-strand breaks.Nucleic Acids Res. 2010 Apr;38(6):1821-31. doi: 10.1093/nar/gkp1164. Epub 2009 Dec 17. Nucleic Acids Res. 2010. PMID: 20019063 Free PMC article.
-
DNA-damage-induced degradation of EXO1 exonuclease limits DNA end resection to ensure accurate DNA repair.J Biol Chem. 2017 Jun 30;292(26):10779-10790. doi: 10.1074/jbc.M116.772475. Epub 2017 May 17. J Biol Chem. 2017. PMID: 28515316 Free PMC article.
-
Regulation of repair pathway choice at two-ended DNA double-strand breaks.Mutat Res. 2017 Oct;803-805:51-55. doi: 10.1016/j.mrfmmm.2017.07.011. Epub 2017 Jul 29. Mutat Res. 2017. PMID: 28781144 Review.
-
Mechanism and regulation of DNA end resection in eukaryotes.Crit Rev Biochem Mol Biol. 2016 May-Jun;51(3):195-212. doi: 10.3109/10409238.2016.1172552. Epub 2016 Apr 20. Crit Rev Biochem Mol Biol. 2016. PMID: 27098756 Free PMC article. Review.
Cited by
-
Targeting neddylation E2s: a novel therapeutic strategy in cancer.J Hematol Oncol. 2021 Apr 7;14(1):57. doi: 10.1186/s13045-021-01070-w. J Hematol Oncol. 2021. PMID: 33827629 Free PMC article. Review.
-
Deficiency of salt-inducible kinase 2 (SIK2) promotes immune injury by inhibiting the maturation of lymphocytes.MedComm (2020). 2023 Sep 11;4(5):e366. doi: 10.1002/mco2.366. eCollection 2023 Oct. MedComm (2020). 2023. PMID: 37706195 Free PMC article.
-
Key molecular DNA damage responses of human cells to radiation.Front Cell Dev Biol. 2024 Jul 10;12:1422520. doi: 10.3389/fcell.2024.1422520. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 39050891 Free PMC article. Review.
-
DNA repair pathway activation features in follicular and papillary thyroid tumors, interrogated using 95 experimental RNA sequencing profiles.Heliyon. 2021 Mar 13;7(3):e06408. doi: 10.1016/j.heliyon.2021.e06408. eCollection 2021 Mar. Heliyon. 2021. PMID: 33748479 Free PMC article.
-
Proteasome-mediated degradation of long-range nucleases negatively regulates resection of DNA double-strand breaks.iScience. 2024 Jun 22;27(7):110373. doi: 10.1016/j.isci.2024.110373. eCollection 2024 Jul 19. iScience. 2024. PMID: 39071887 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

