Horizontal acquisition of a patchwork Calvin cycle by symbiotic and free-living Campylobacterota (formerly Epsilonproteobacteria)
- PMID: 31562384
- PMCID: PMC6908604
- DOI: 10.1038/s41396-019-0508-7
Horizontal acquisition of a patchwork Calvin cycle by symbiotic and free-living Campylobacterota (formerly Epsilonproteobacteria)
Abstract
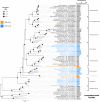
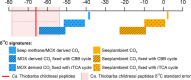
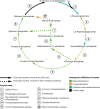
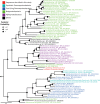
Most autotrophs use the Calvin-Benson-Bassham (CBB) cycle for carbon fixation. In contrast, all currently described autotrophs from the Campylobacterota (previously Epsilonproteobacteria) use the reductive tricarboxylic acid cycle (rTCA) instead. We discovered campylobacterotal epibionts ("Candidatus Thiobarba") of deep-sea mussels that have acquired a complete CBB cycle and may have lost most key genes of the rTCA cycle. Intriguingly, the phylogenies of campylobacterotal CBB cycle genes suggest they were acquired in multiple transfers from Gammaproteobacteria closely related to sulfur-oxidizing endosymbionts associated with the mussels, as well as from Betaproteobacteria. We hypothesize that "Ca. Thiobarba" switched from the rTCA cycle to a fully functional CBB cycle during its evolution, by acquiring genes from multiple sources, including co-occurring symbionts. We also found key CBB cycle genes in free-living Campylobacterota, suggesting that the CBB cycle may be more widespread in this phylum than previously known. Metatranscriptomics and metaproteomics confirmed high expression of CBB cycle genes in mussel-associated "Ca. Thiobarba". Direct stable isotope fingerprinting showed that "Ca. Thiobarba" has typical CBB signatures, suggesting that it uses this cycle for carbon fixation. Our discovery calls into question current assumptions about the distribution of carbon fixation pathways in microbial lineages, and the interpretation of stable isotope measurements in the environment.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Rosgaard L, de Porcellinis AJ, Jacobsen JH, Frigaard N-U, Sakuragi Y. Bioengineering of carbon fixation, biofuels, and biochemicals in cyanobacteria and plants. J Biotechnol. 2012;162:134–47. - PubMed
-
- Hügler M, Sievert SM. Beyond the Calvin cycle: autotrophic carbon fixation in the ocean. Ann Rev Mar Sci. 2011;3:261–89. - PubMed
-
- Raven J. Contributions of anoxygenic and oxygenic phototrophy and chemolithotrophy to carbon and oxygen fluxes in aquatic environments. Aquat Micro Ecol. 2009;56:177–92.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

