Circular RNA profiling in the oocyte and cumulus cells reveals that circARMC4 is essential for porcine oocyte maturation
- PMID: 31562810
- PMCID: PMC6781969
- DOI: 10.18632/aging.102315
Circular RNA profiling in the oocyte and cumulus cells reveals that circARMC4 is essential for porcine oocyte maturation
Abstract
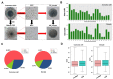
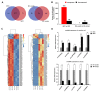
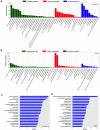
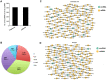
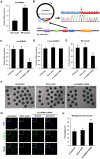
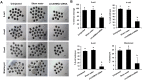
Thousands of circular RNAs (circRNAs) have been recently discovered in cumulus cells and oocytes from several species. However, the expression and function of circRNA during porcine oocyte meiotic maturation have been never examined. Here, we separately identified 7,067 and 637 circRNAs in both cumulus cells and oocytes via deep sequencing and bioinformatic analysis. Further analysis revealed that a faction of circRNAs is differentially expressed (DE) in a developmental stage-specific manner. The host genes of DE circRNAs are markedly enriched to multiple signaling pathways associated with cumulus cell function and oocyte maturation. Additionally, most DE circRNAs harbor several miRNA targets, suggesting that these DE circRNAs potentially act as miRNA sponge. Importantly, we found that maternal circARMC4 knockdown by siRNA microinjection caused a severely impaired chromosome alignment, and significantly inhibited first polar body extrusion and early embryo development. Taken together, these results demonstrate for the first time that circRNAs are abundantly and dynamically expressed in a developmental stage-specific manner in cumulus cells and oocytes, and maternally expressed circARMC4 is essential for porcine oocyte meiotic maturation and early embryo development.
Keywords: circular RNA; cumulus cells; meiotic maturation; oocytes; pig.
Conflict of interest statement
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

