The spectrum of APOBEC3 activity: From anti-viral agents to anti-cancer opportunities
- PMID: 31563041
- PMCID: PMC6876854
- DOI: 10.1016/j.dnarep.2019.102700
The spectrum of APOBEC3 activity: From anti-viral agents to anti-cancer opportunities
Abstract
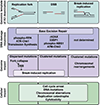
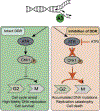
The APOBEC3 family of cytosine deaminases are part of the innate immune response to viral infection, but also have the capacity to damage cellular DNA. Detection of mutational signatures consistent with APOBEC3 activity, together with elevated APOBEC3 expression in cancer cells, has raised the possibility that these enzymes contribute to oncogenesis. Genome deamination by APOBEC3 enzymes also elicits DNA damage response signaling and presents therapeutic vulnerabilities for cancer cells. Here, we discuss implications of APOBEC3 activity in cancer and the potential to exploit their mutagenic activity for targeted cancer therapies.
Keywords: APOBEC3; Cytosine deaminase; DNA damage response; Mutational patterns; Synthetic lethality.
Copyright © 2019 Elsevier B.V. All rights reserved.
Figures

References
-
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N, An anthropoid-specific locus of orphan C to U RNA-editing enzymes on chromosome 22, Genomics, 79 (2002) 285–296. - PubMed
-
- Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T, Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme, Cell, 102 (2000) 553–563. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

