Investigating Transfusion-related Sepsis Using Culture-Independent Metagenomic Sequencing
- PMID: 31563940
- PMCID: PMC7442849
- DOI: 10.1093/cid/ciz960
Investigating Transfusion-related Sepsis Using Culture-Independent Metagenomic Sequencing
Abstract
Background: Transfusion-related sepsis remains an important hospital infection control challenge. Investigation of septic transfusion events is often restricted by the limitations of bacterial culture in terms of time requirements and low yield in the setting of prior antibiotic administration.
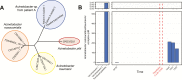
Methods: In 3 gram-negative septic transfusion cases, we performed metagenomic next-generation sequencing (mNGS) of direct clinical blood specimens in addition to standard culture-based approaches utilized for infection control investigations. Pathogen detection leveraged IDSeq, a new open-access microbial bioinformatics portal. Phylogenetic analysis was performed to assess microbial genetic relatedness and understand transmission events.
Results: mNGS of direct clinical blood specimens afforded precision detection of pathogens responsible for each case of transfusion-related sepsis and enabled discovery of a novel Acinetobacter species in a platelet product that had become contaminated despite photochemical pathogen reduction. In each case, longitudinal assessment of pathogen burden elucidated the temporal sequence of events associated with each transfusion-transmitted infection. We found that informative data could be obtained from culture-independent mNGS of residual platelet products and leftover blood specimens that were either unsuitable or unavailable for culture or that failed to grow due to prior antibiotic administration. We additionally developed methods to enhance accuracy for detecting transfusion-associated pathogens that share taxonomic similarity to contaminants commonly found in mNGS library preparations.
Conclusions: Culture-independent mNGS of blood products afforded rapid and precise assessment of pathogen identity, abundance, and genetic relatedness. Together, these challenging cases demonstrated the potential for metagenomics to advance existing methods for investigating transfusion-transmitted infections.
Keywords: healthcare infections; mNGS; metagenomic sequencing; platelet transfusion; septic transfusion.
© The Author(s) 2019. Published by Oxford University Press for the Infectious Diseases Society of America.
Figures


References
-
- Eder AF, Dy BA, DeMerse B, et al. . Apheresis technology correlates with bacterial contamination of platelets and reported septic transfusion reactions: : Apheresis Technology and Bacterial Contamination of PLTs. Transfusion 2017; 57:2969–76. - PubMed
-
- Busch MP, Bloch EM, Kleinman S. Prevention of transfusion-transmitted infections. Blood 2019; 133:1854–64. - PubMed
-
- Tormey CA, Sweeney JD, Champion MH, Pisciotto PT, Snyder EL, Wu Y. Analysis of transfusion reactions associated with prestorage-pooled platelet components. Transfusion 2009; 49:1242–7. - PubMed
-
- Kuehnert MJ, Roth VR, Haley NR, et al. . Transfusion-transmitted bacterial infection in the United States, 1998 through 2000. Transfusion 2001; 41:1493–9. - PubMed

