Harmonizing Genetic Ancestry and Self-identified Race/Ethnicity in Genome-wide Association Studies
- PMID: 31564439
- PMCID: PMC6817526
- DOI: 10.1016/j.ajhg.2019.08.012
Harmonizing Genetic Ancestry and Self-identified Race/Ethnicity in Genome-wide Association Studies
Abstract
Large-scale multi-ethnic cohorts offer unprecedented opportunities to elucidate the genetic factors influencing complex traits related to health and disease among minority populations. At the same time, the genetic diversity in these cohorts presents new challenges for analysis and interpretation. We consider the utility of race and/or ethnicity categories in genome-wide association studies (GWASs) of multi-ethnic cohorts. We demonstrate that race/ethnicity information enhances the ability to understand population-specific genetic architecture. To address the practical issue that self-identified racial/ethnic information may be incomplete, we propose a machine learning algorithm that produces a surrogate variable, termed HARE. We use height as a model trait to demonstrate the utility of HARE and ethnicity-specific GWASs.
Keywords: biobank; ethnicity-specific trait loci; genetic ancestry; multi-ethnic cohort; self-reported race/ethnicity; stratified analysis; trans-ethnic GWAS.
Copyright © 2019 American Society of Human Genetics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
S.L.D. has received research grants from the following for-profit organizations in the last three years: AbbVie Inc., Anolinx LLC, Astellas Pharma Inc., AstraZeneca Pharmaceuticals LP, Boehringer Ingelheim International GmbH, Celgene Corporation, Eli Lilly and Company, Genentech Inc., Genomic Health, Inc., Gilead Sciences Inc., GlaxoSmithKline PLC, Innocrin Pharmaceuticals Inc., Janssen Pharmaceuticals, Inc., Kantar Health, Myriad Genetic Laboratories, Inc., Novartis International AG, and PAREXEL International Corporation through the University of Utah or Western Institute for Biomedical Research. S.M.D. has received research grant from RenalytixAI and CytoVas through the University of Pennsylvania.
Figures
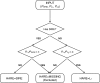
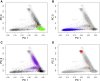
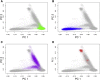
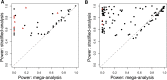
References
-
- Gaziano J.M., Concato J., Brophy M., Fiore L., Pyarajan S., Breeling J., Whitbourne S., Deen J., Shannon C., Humphries D. Million Veteran Program: A mega-biobank to study genetic influences on health and disease. J. Clin. Epidemiol. 2016;70:214–223. - PubMed
-
- Tang H., Peng J., Wang P., Risch N.J. Estimation of individual admixture: analytical and study design considerations. Genet. Epidemiol. 2005;28:289–301. - PubMed
-
- Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. - PubMed
-
- Coram M.A., Duan Q., Hoffmann T.J., Thornton T., Knowles J.W., Johnson N.A., Ochs-Balcom H.M., Donlon T.A., Martin L.W., Eaton C.B. Genome-wide characterization of shared and distinct genetic components that influence blood lipid levels in ethnically diverse human populations. Am. J. Hum. Genet. 2013;92:904–916. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

