Reactivity of N3-Methyl-2'-Deoxyadenosine in Nucleosome Core Particles
- PMID: 31565933
- PMCID: PMC6803048
- DOI: 10.1021/acs.chemrestox.9b00299
Reactivity of N3-Methyl-2'-Deoxyadenosine in Nucleosome Core Particles
Abstract
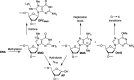
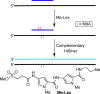
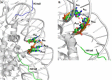
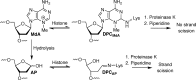
N3-Methyl-2'-deoxyadenosine (MdA) is the major dA methylation product in duplex DNA. MdA blocks DNA replication and undergoes depurination at significantly higher rates than the native nucleotide from which it is derived. Recent reports on the effects of the nucleosome core particle (NCP) environment on the reactivity of N7-methyl-2'-deoxyguanosine (MdG) inspired this investigation concerning the reactivity of MdA in NCPs. NCPs containing MdA at selected positions were produced using a strategy in which the minor groove binding Me-Lex molecule serves as a sequence specific methylating agent. Hydrolysis of the glycosidic bond in MdA to form abasic sites (AP) is suppressed in a NCP. Experiments using histone variants indicate that the proximal, highly basic N-terminal tails are partially responsible for the decreased depurination rate constant. MdA also forms cross-links with histone proteins. The levels of MdA-histone DNA-protein cross-links (DPCMdA) decrease significantly over time and are replaced by those involving AP. The time dependent decrease in DPCMdA is attributed to the reversibility of their formation and the relatively rapid rate of AP formation from MdA. Overall, MdA reactivity in NCPs qualitatively resembles that of MdG.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

