Early adaptive immune activation detected in monozygotic twins with prodromal multiple sclerosis
- PMID: 31566584
- PMCID: PMC6819125
- DOI: 10.1172/JCI128475
Early adaptive immune activation detected in monozygotic twins with prodromal multiple sclerosis
Abstract
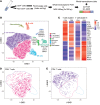
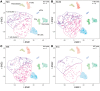
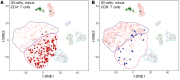
Multiple sclerosis (MS) is a disabling disease of the CNS. Inflammatory features of MS include lymphocyte accumulations in the CNS and cerebrospinal fluid (CSF). The preclinical events leading to established MS are still enigmatic. Here we compared gene expression patterns of CSF cells from MS-discordant monozygotic twin pairs. Six "healthy" co-twins, who carry a maximal familial risk for developing MS, showed subclinical neuroinflammation (SCNI) with small MRI lesions. Four of these subjects had oligoclonal bands (OCBs). By single-cell RNA sequencing of 2752 CSF cells, we identified clonally expanded CD8+ T cells, plasmablasts, and, to a lesser extent, CD4+ T cells not only from MS patients but also from subjects with SCNI. In contrast to nonexpanded T cells, clonally expanded T cells showed characteristics of activated tissue-resident memory T (TRM) cells. The TRM-like phenotype was detectable already in cells from SCNI subjects but more pronounced in cells from patients with definite MS. Expanded plasmablast clones were detected only in MS and SCNI subjects with OCBs. Our data provide evidence for very early concomitant activation of 3 components of the adaptive immune system in MS, with a notable contribution of clonally expanded TRM-like CD8+ cells.
Keywords: Adaptive immunity; Immunology; Multiple sclerosis; Neuroscience.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

