Breakpoint delineation in 5p- patients leads to new insights about microcephaly and the typical high-pitched cry
- PMID: 31568707
- PMCID: PMC7005617
- DOI: 10.1002/mgg3.957
Breakpoint delineation in 5p- patients leads to new insights about microcephaly and the typical high-pitched cry
Abstract
Background: Cri du chat syndrome (CdCS) is a rare syndrome caused by a partial or complete deletion of the short arm of chromosome 5 (5p-). The main clinical features include a high-pitched cry, facial asymmetry, microcephaly, round face at birth, epicanthal folds, hypotonia, delayed growth and development.
Methods: We studied 14 Brazilian patients with CdCS using genomic array in order to better define the 5p breakpoints and recognize copy number variations (CNVs) that contribute to clinical manifestations associated with the syndrome.
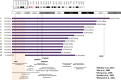
Results: Array confirmed terminal deletions in 13 patients and an interstitial deletion in one patient. It was also possible to map the breakpoints and associate a genomic region of 4.7 Mb to the development of head circumference and cat-like cry. We also found other CNVs concomitant to the 5p deletion including a 9p duplication, a 17q deletion, and a 22q deletion in three different patients.
Conclusion: With advancements of molecular cytogenomic methods in the last two decades, it was possible to evidence cryptic alterations and improve the genotype-phenotype correlation. In this work, we describe a new genomic region associated with microcephaly and cat-like cry and highlight the importance of precise delineation of 5p deletion breakpoints and detection of other CNVs in CdCS patients to improve genotype-phenotype correlation to perform a complete clinical and molecular diagnosis.
Keywords: Brazilian patients; cri du chat; cytogenomic; genomic array.
© 2019 The Authors. Molecular Genetics & Genomic Medicine published by Wiley Periodicals, Inc.
Conflict of interest statement
There are no conflict of interest to disclose.
Figures

References
-
- Ballif, B. C. , Wakui, K. , Gajecka, M. , & Shaffer, L. G. (2004). Translocation breakpoint mapping and sequence analysis in three monosomy 1p36 subjects with der(1)t(1;1)(p36;q44) suggest mechanisms for telomere capture in stabilizing de novo terminal rearrangements. Human Genetics, 114(2), 198–206. 10.1007/s00439-003-1029-y - DOI - PubMed
-
- Elmakky, A. , Carli, D. , Lugli, L. , Torelli, P. , Guidi, B. , Falcinelli, C. , … Percesepe, A. (2014). A three‐generation family with terminal microdeletion involving 5p15.33‐32 due to a whole‐arm 5;15 chromosomal translocation with a steady phenotype of atypical cri du chat syndrome. European Journal of Medical Genetics, 57(4), 145–150. 10.1016/j.ejmg.2014.02.005 - DOI - PubMed
-
- Kearney, H. M. , Thorland, E. C. , Brown, K. K. , Quintero‐Rivera, F. , & South, S. T. ; Working Group of the American College of Medical Genetics Laboratory Quality Assurance Committee . (2011). American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genetics in Medicine, 13(7), 680–685. 10.1097/GIM.0b013e3182217a3a - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

