Membrane Topological Model of Glycosyltransferases of the GT-C Superfamily
- PMID: 31569500
- PMCID: PMC6801728
- DOI: 10.3390/ijms20194842
Membrane Topological Model of Glycosyltransferases of the GT-C Superfamily
Abstract
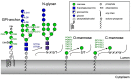
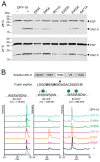
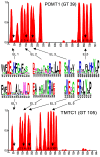
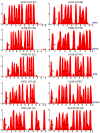
Glycosyltransferases that use polyisoprenol-linked donor substrates are categorized in the GT-C superfamily. In eukaryotes, they act in the endoplasmic reticulum (ER) lumen and are involved in N-glycosylation, glypiation, O-mannosylation, and C-mannosylation of proteins. We generated a membrane topology model of C-mannosyltransferases (DPY19 family) that concurred perfectly with the 13 transmembrane domains (TMDs) observed in oligosaccharyltransferases (STT3 family) structures. A multiple alignment of family members from diverse organisms highlighted the presence of only a few conserved amino acids between DPY19s and STT3s. Most of these residues were shown to be essential for DPY19 function and are positioned in luminal loops that showed high conservation within the DPY19 family. Multiple alignments of other eukaryotic GT-C families underlined the presence of similar conserved motifs in luminal loops, in all enzymes of the superfamily. Most GT-C enzymes are proposed to have an uneven number of TDMs with 11 (POMT, TMTC, ALG9, ALG12, PIGB, PIGV, and PIGZ) or 13 (DPY19, STT3, and ALG10) membrane-spanning helices. In contrast, PIGM, ALG3, ALG6, and ALG8 have 12 or 14 TMDs and display a C-terminal dilysine ER-retrieval motif oriented towards the cytoplasm. We propose that all members of the GT-C superfamily are evolutionary related enzymes with preserved membrane topology.
Keywords: C-mannosylation; GPI-anchor; N-glycosylation; O-mannosylation; dolichol-phosphate; endoplasmic reticulum; glycosyltransferase; mannose; mannosyltransferase; oligosaccharyltransferase.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
-
- Larsen I.S.B., Narimatsu Y., Joshi H.J., Siukstaite L., Harrison O.J., Brasch J., Goodman K.M., Hansen L., Shapiro L., Honig B., et al. Discovery of an O-mannosylation pathway selectively serving cadherins and protocadherins. Proc. Natl. Acad. Sci. USA. 2017;114:11163–11168. doi: 10.1073/pnas.1708319114. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

