SSEThread: Integrative threading of the DNA-PKcs sequence based on data from chemical cross-linking and hydrogen deuterium exchange
- PMID: 31570166
- PMCID: PMC6903780
- DOI: 10.1016/j.pbiomolbio.2019.09.003
SSEThread: Integrative threading of the DNA-PKcs sequence based on data from chemical cross-linking and hydrogen deuterium exchange
Abstract
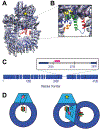
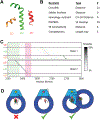
X-ray crystallography and electron microscopy maps resolved to 3-8 Å are generally sufficient for tracing the path of the polypeptide chain in space, while often insufficient for unambiguously registering the sequence on the path (i.e., threading). Frequently, however, additional information is available from other biophysical experiments, physical principles, statistical analyses, and other prior models. Here, we formulate an integrative approach for sequence assignment to a partial backbone model as an optimization problem, which requires three main components: the representation of the system, the scoring function, and the optimization method. The method is implemented in the open source Integrative Modeling Platform (IMP) (https://integrativemodeling.org), allowing a number of different terms in the scoring function. We apply this method to localizing the sequence assignment within a 199-residue disordered region of three structured and sequence unassigned helices in the DNA-PKcs crystallographic structure, using chemical crosslinks, hydrogen deuterium exchange, and sequence connectivity. The resulting ensemble of threading models provides two major solutions, one of which suggests that the crucial ABCDE cluster of phosphorylation sites cannot undergo intra-molecular autophosphorylation without a conformational rearrangement. The ensemble of solutions embodies the most accurate and precise sequence threading given the available information.
Keywords: DNA-PKcs; Electron microscopy; Integrative modeling; Threading; X-ray crystallography.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Figures




References
-
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Rout MP, Sali A, 2007. Determining the architectures of macromolecular assemblies. Nature 450, 683–694. - PubMed
-
- Alber F, Förster F, Korkin D, Topf M, Sali A, 2008. Integrating diverse data for structure determination of macromolecular assemblies. Annu Rev Biochem 77, 443–477. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

