Tripartite Regulation of the glpFKD Operon Involved in Glycerol Catabolism by GylR, Crp, and SigF in Mycobacterium smegmatis
- PMID: 31570530
- PMCID: PMC6872199
- DOI: 10.1128/JB.00511-19
Tripartite Regulation of the glpFKD Operon Involved in Glycerol Catabolism by GylR, Crp, and SigF in Mycobacterium smegmatis
Abstract
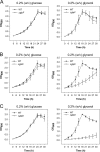
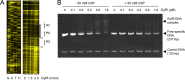
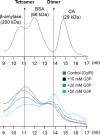
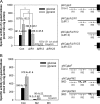
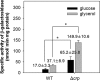
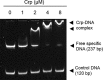
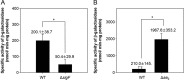
The glpD (MSMEG_6761) gene encoding glycerol-3-phosphate dehydrogenase was shown to be crucial for M. smegmatis to utilize glycerol as the sole carbon source. The glpD gene likely forms the glpFKD operon together with glpF and glpK, encoding a glycerol facilitator and glycerol kinase, respectively. The gylR (MSMEG_6757) gene, whose product belongs to the IclR family of transcriptional regulators, was identified 182 bp upstream of glpF It was demonstrated that GylR serves as a transcriptional activator and is involved in the induction of glpFKD expression in the presence of glycerol. Three GylR-binding sites with the consensus sequence (GKTCGRC-N3-GYCGAMC) were identified in the upstream region of glpF by DNase I footprinting analysis. The presence of glycerol-3-phosphate was shown to decrease the binding affinity of GylR to the glpF upstream region with changes in the quaternary structure of GylR from tetramer to dimer. Besides GylR, cAMP receptor protein (Crp) and an alternative sigma factor, SigF, are also implicated in the regulation of glpFKD expression. Crp functions as a repressor, while SigF induces expression of glpFKD under energy-limiting conditions. In conclusion, we suggest here that the glpFKD operon is under the tripartite control of GylR, SigF, and Crp, which enables M. smegmatis to integrate the availability of glycerol, cellular energy state, and cellular levels of cAMP to exquisitely control expression of the glpFKD operon involved in glycerol metabolism.IMPORTANCE Using genetic approaches, we first revealed that glycerol is catabolized through the glycolytic pathway after conversion to dihydroxyacetone phosphate in two sequential reactions catalyzed by glycerol kinase (GlpK) and flavin adenine dinucleotide (FAD)-containing glycerol-3-phosphate dehydrogenase (GlpD) in M. smegmatis Our study also revealed that in addition to the GylR transcriptional activator that mediates the induction of the glpFKD operon by glycerol, the operon is regulated by SigF and Crp, which reflect the cellular energy state and cAMP level, respectively.
Keywords: GylR transcriptional regulator; Mycobacterium; SigF; cAMP receptor protein; glycerol catabolism; glycerol kinase; glycerol-3-phosphate dehydrogenase; regulation of gene expression.
Copyright © 2019 American Society for Microbiology.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

