How internal cavities destabilize a protein
- PMID: 31570587
- PMCID: PMC6800337
- DOI: 10.1073/pnas.1911181116
How internal cavities destabilize a protein
Abstract
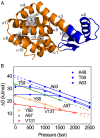
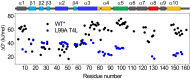
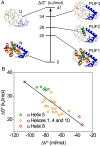
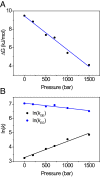
Although many proteins possess a distinct folded structure lying at a minimum in a funneled free energy landscape, thermal energy causes any protein to continuously access lowly populated excited states. The existence of excited states is an integral part of biological function. Although transitions into the excited states may lead to protein misfolding and aggregation, little structural information is currently available for them. Here, we show how NMR spectroscopy, coupled with pressure perturbation, brings these elusive species to light. As pressure acts to favor states with lower partial molar volume, NMR follows the ensuing change in the equilibrium spectroscopically, with residue-specific resolution. For T4 lysozyme L99A, relaxation dispersion NMR was used to follow the increase in population of a previously identified "invisible" folded state with pressure, as this is driven by the reduction in cavity volume by the flipping-in of a surface aromatic group. Furthermore, multiple partly disordered excited states were detected at equilibrium using pressure-dependent H/D exchange NMR spectroscopy. Here, unfolding reduced partial molar volume by the removal of empty internal cavities and packing imperfections through subglobal and global unfolding. A close correspondence was found for the distinct pressure sensitivities of various parts of the protein and the amount of internal cavity volume that was lost in each unfolding event. The free energies and populations of excited states allowed us to determine the energetic penalty of empty internal protein cavities to be 36 cal⋅Å-3.
Keywords: high-pressure NMR; protein folding and cooperativity; protein stability; unfolded state.
Conflict of interest statement
The authors declare no competing interest.
Figures
References
-
- Nicholls A., Sharp K. A., Honig B., Protein folding and association: Insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins 11, 281–296 (1991). - PubMed
-
- Dobson C. M., Protein folding and misfolding. Nature 426, 884–890 (2003). - PubMed
-
- Chiti F., Dobson C. M., Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 (2017). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

