Inherent reactivity of unselected TCR repertoires to peptide-MHC molecules
- PMID: 31570608
- PMCID: PMC6825295
- DOI: 10.1073/pnas.1909504116
Inherent reactivity of unselected TCR repertoires to peptide-MHC molecules
Abstract
The repertoire of αβ T cell antigen receptors (TCRs) on mature T cells is selected in the thymus where it is rendered both self-tolerant and restricted to the recognition of major histocompatibility complex molecules presenting peptide antigens (pMHC). It remains unclear whether germline TCR sequences exhibit an inherent bias to interact with pMHC prior to selection. Here, we isolated TCR libraries from unselected thymocytes and upon reexpression of these random TCR repertoires in recipient T cell hybridomas, interrogated their reactivities to antigen-presenting cell lines. While these random TCR combinations could potentially have reacted with any surface molecule on the cell lines, the hybridomas were stimulated most frequently by pMHC ligands. The nature and CDR3 loop composition of the TCRβ chain played a dominant role in determining pMHC-reactivity. Replacing the germline regions of mouse TCRβ chains with those of other jawed vertebrates preserved reactivity to mouse pMHC. Finally, introducing the CD4 coreceptor into the hybridomas increased the proportion of cells that could respond to pMHC ligands. Thus, αβ TCRs display an intrinsic and evolutionary conserved bias for pMHC molecules in the absence of any selective pressure, which is further strengthened in the presence of coreceptors.
Keywords: T cell antigen receptor; T cell selection; major histocompatibility complex.
Copyright © 2019 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no competing interest.
Figures

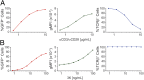

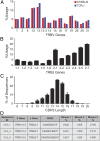



Comment in
-
T cell antigen recognition: Evolution-driven affinities.Proc Natl Acad Sci U S A. 2019 Oct 29;116(44):21969-21971. doi: 10.1073/pnas.1916129116. Epub 2019 Oct 14. Proc Natl Acad Sci U S A. 2019. PMID: 31611377 Free PMC article. No abstract available.
References
-
- Schatz D. G., Ji Y., Recombination centres and the orchestration of V(D)J recombination. Nat. Rev. Immunol. 11, 251–263 (2011). - PubMed
-
- Davis M. M., Bjorkman P. J., T-cell antigen receptor genes and T-cell recognition. Nature 334, 395–402 (1988). - PubMed
-
- Rudolph M. G., Stanfield R. L., Wilson I. A., How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24, 419–466 (2006). - PubMed
-
- La Gruta N. L., Gras S., Daley S. R., Thomas P. G., Rossjohn J., Understanding the drivers of MHC restriction of T cell receptors. Nat. Rev. Immunol. 18, 467–478 (2018). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

