X chromosome and autosomal recombination are differentially sensitive to disruptions in SC maintenance
- PMID: 31570610
- PMCID: PMC6815145
- DOI: 10.1073/pnas.1910840116
X chromosome and autosomal recombination are differentially sensitive to disruptions in SC maintenance
Abstract
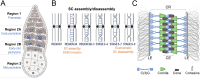
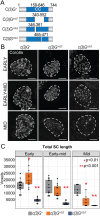
The synaptonemal complex (SC) is a conserved meiotic structure that regulates the repair of double-strand breaks (DSBs) into crossovers or gene conversions. The removal of any central-region SC component, such as the Drosophila melanogaster transverse filament protein C(3)G, causes a complete loss of SC structure and crossovers. To better understand the role of the SC in meiosis, we used CRISPR/Cas9 to construct 3 in-frame deletions within the predicted coiled-coil region of the C(3)G protein. Since these 3 deletion mutations disrupt SC maintenance at different times during pachytene and exhibit distinct defects in key meiotic processes, they allow us to define the stages of pachytene when the SC is necessary for homolog pairing and recombination during pachytene. Our studies demonstrate that the X chromosome and the autosomes display substantially different defects in pairing and recombination when SC structure is disrupted, suggesting that the X chromosome is potentially regulated differently from the autosomes.
Keywords: Drosophila; homologous recombination; meiosis; synaptonemal complex.
Conflict of interest statement
The authors declare no competing interest.
Figures





References
-
- Hassold T., Hall H., Hunt P., The origin of human aneuploidy: Where we have been, where we are going. Hum. Mol. Genet. 16, R203–R208 (2007). - PubMed
-
- Keeney S., Giroux C. N., Kleckner N., Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88, 375–384 (1997). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

