Targeted isolation and cultivation of uncultivated bacteria by reverse genomics
- PMID: 31570900
- PMCID: PMC6858544
- DOI: 10.1038/s41587-019-0260-6
Targeted isolation and cultivation of uncultivated bacteria by reverse genomics
Abstract
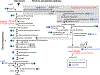
Most microorganisms from all taxonomic levels are uncultured. Single-cell genomes and metagenomes continue to increase the known diversity of Bacteria and Archaea; however, while 'omics can be used to infer physiological or ecological roles for species in a community, most of these hypothetical roles remain unvalidated. Here, we report an approach to capture specific microorganisms from complex communities into pure cultures using genome-informed antibody engineering. We apply our reverse genomics approach to isolate and sequence single cells and to cultivate three different species-level lineages of human oral Saccharibacteria (TM7). Using our pure cultures, we show that all three Saccharibacteria species are epibionts of diverse Actinobacteria. We also isolate and cultivate human oral SR1 bacteria, which are members of a lineage of previously uncultured bacteria. Reverse-genomics-enabled cultivation of microorganisms can be applied to any species from any environment and has the potential to unlock the isolation, cultivation and characterization of species from as-yet-uncultured branches of the microbial tree of life.
Conflict of interest statement
Figures
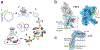


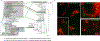

Comment in
-
Culturing the uncultured.Nat Biotechnol. 2019 Nov;37(11):1278-1279. doi: 10.1038/s41587-019-0300-2. Nat Biotechnol. 2019. PMID: 31597966 No abstract available.
-
Culturing uncultivated bacteria.Nat Methods. 2019 Nov;16(11):1078. doi: 10.1038/s41592-019-0634-1. Nat Methods. 2019. PMID: 31673145 No abstract available.
References
-
- DeLong EF & Pace NR Environmental diversity of bacteria and archaea. Syst Biol 50, 470–478 (2001). - PubMed
-
- Hug LA et al. A new view of the tree of life. Nature microbiology 1, 16048 (2016). - PubMed
-
- Zaremba-Niedzwiedzka K et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541, 353–358 (2017). - PubMed
-
- Parks DH et al. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nature microbiology 2, 1533–1542 (2017). - PubMed
-
- Rinke C et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499, 431–437 (2013). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

