Can Insertion Sequences Proliferation Influence Genomic Plasticity? Comparative Analysis of Acinetobacter baumannii Sequence Type 78, a Persistent Clone in Italian Hospitals
- PMID: 31572316
- PMCID: PMC6751323
- DOI: 10.3389/fmicb.2019.02080
Can Insertion Sequences Proliferation Influence Genomic Plasticity? Comparative Analysis of Acinetobacter baumannii Sequence Type 78, a Persistent Clone in Italian Hospitals
Abstract
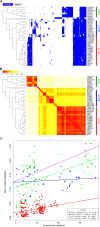
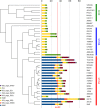
Acinetobacter baumannii is a known opportunistic pathogen, dangerous for public health mostly due to its ability to rapidly acquire antibiotic-resistance traits. Its genome was described as characterized by remarkable plasticity, with a high frequency of homologous recombinations and proliferation of Insertion Sequences (IS). The SMAL pulsotype is an A. baumannii strain currently isolated only in Italy, characterized by a low incidence and a high persistence over the years. In this present work, we have conducted a comparative genomic analysis on this clone. The genome of 15 SMAL isolates was obtained and characterized in comparison with 24 other assemblies of evolutionary related isolates. The phylogeny highlighted the presence of a monophyletic clade (named ST78A), which includes the SMAL isolates. ST78A isolates have a low rate of homologous recombination and low gene content variability when compared to two related clades (ST78B and ST49) and to the most common A. baumannii variants worldwide (International Clones I and II). Remarkably, genomes in the ST78A clade present a high number of IS, including classes mostly absent in the other related genomes. Among these IS, one copy of IS66 was found to interrupt the gene comEC/rec2, involved in the acquisition of exogenous DNA. The genomic characterization of SMAL isolates shed light on the surprisingly low genomic plasticity and the high IS proliferation present in this strain. The interruption of the gene comEC/rec2 by an IS in the SMAL genomes brought us to formulate an evolutionary hypothesis according to which the proliferation of IS is slowing the acquisition of exogenous DNA, thus limiting genome plasticity. Such genomic architecture could explain the epidemiological behavior of high persistence and low incidence of the clone and provides an interesting framework to compare ST78 with the highly epidemic International Clones, characterized by high genomic plasticity.
Keywords: Acinetobacter baumannii; SMAL; ST78; genomic plasticity; insertion sequences.
Copyright © 2019 Gaiarsa, Bitar, Comandatore, Corbella, Piazza, Scaltriti, Villa, Postiglione, Marone, Nucleo, Pongolini, Migliavacca and Sassera.
Figures

References
-
- Beare P. A., Unsworth N., Andoh M., Voth D. E., Omsland A., Gilk S. D., et al. (2009). Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect. Immun. 77 642–656. 10.1128/IAI.01141-08 - DOI - PMC - PubMed

