Large-Scale Analysis of the Mycoplasma bovis Genome Identified Non-essential, Adhesion- and Virulence-Related Genes
- PMID: 31572317
- PMCID: PMC6753880
- DOI: 10.3389/fmicb.2019.02085
Large-Scale Analysis of the Mycoplasma bovis Genome Identified Non-essential, Adhesion- and Virulence-Related Genes
Abstract
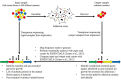
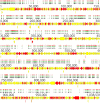
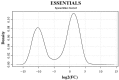
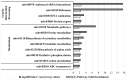
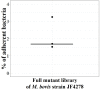
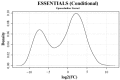
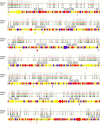
Mycoplasma bovis is an important pathogen of cattle causing bovine mycoplasmosis. Clinical manifestations are numerous, but pneumonia, mastitis, and arthritis cases are mainly reported. Currently, no efficient vaccine is available and antibiotic treatments are not always satisfactory. The design of new, efficient prophylactic and therapeutic approaches requires a better understanding of the molecular mechanisms responsible for M. bovis pathogenicity. Random transposon mutagenesis has been widely used in Mycoplasma species to identify potential gene functions. Such an approach can also be used to screen genomes and search for essential and non-essential genes for growth. Here, we generated a random transposon mutant library of M. bovis strain JF4278 containing approximately 4000 independent insertion sites. We then coupled high-throughput screening of this mutant library to transposon sequencing and bioinformatic analysis to identify M. bovis non-essential, adhesion- and virulence-related genes. Three hundred and fifty-two genes of M. bovis were assigned as essential for growth in rich medium. Among the remaining non-essential genes, putative virulence-related factors were subsequently identified. The complete mutant library was screened for adhesion using primary bovine mammary gland epithelial cells. Data from this assay resulted in a list of conditional-essential genes with putative adhesion-related functions by identifying non-essential genes for growth that are essential for host cell-adhesion. By individually assessing the adhesion capacity of six selected mutants, two previously unknown factors and the adhesin TrmFO were associated with a reduced adhesion phenotype. Overall, our study (i) uncovers new, putative virulence-related genes; (ii) offers a list of putative adhesion-related factors; and (iii) provides valuable information for vaccine design and for exploring M. bovis biology, pathogenesis, and host-interaction.
Keywords: Mycoplasma bovis; adhesion; non-essential genes; random transposon mutagenesis; virulence.
Copyright © 2019 Josi, Bürki, Vidal, Dordet-Frisoni, Citti, Falquet and Pilo.
Figures

Similar articles
-
Bovine Epithelial in vitro Infection Models for Mycoplasma bovis.Front Cell Infect Microbiol. 2018 Sep 18;8:329. doi: 10.3389/fcimb.2018.00329. eCollection 2018. Front Cell Infect Microbiol. 2018. PMID: 30280094 Free PMC article.
-
Genes required for survival and proliferation of Mycoplasma bovis in association with host cells.Appl Environ Microbiol. 2024 Jul 24;90(7):e0068724. doi: 10.1128/aem.00687-24. Epub 2024 Jun 12. Appl Environ Microbiol. 2024. PMID: 38864628 Free PMC article.
-
The presence of adhesion factors NOX, α-enolase, TrmFO, P27, and VpmaX in Mycoplasma bovis wild isolates in Japan.Open Vet J. 2022 Nov-Dec;12(6):782-786. doi: 10.5455/OVJ.2022.v12.i6.1. Epub 2022 Nov 3. Open Vet J. 2022. PMID: 36650870 Free PMC article.
-
Virulence, persistence and dissemination of Mycoplasma bovis.Vet Microbiol. 2015 Aug 31;179(1-2):15-22. doi: 10.1016/j.vetmic.2015.02.024. Epub 2015 Mar 2. Vet Microbiol. 2015. PMID: 25824130 Review.
-
Proteomics analysis and its role in elucidation of functionally significant proteins in Mycoplasma bovis.Microb Pathog. 2017 Oct;111:50-59. doi: 10.1016/j.micpath.2017.08.024. Epub 2017 Aug 18. Microb Pathog. 2017. PMID: 28826762 Review.
Cited by
-
An emerging role for cyclic dinucleotide phosphodiesterase and nanoRNase activities in Mycoplasma bovis: Securing survival in cell culture.PLoS Pathog. 2020 Jun 29;16(6):e1008661. doi: 10.1371/journal.ppat.1008661. eCollection 2020 Jun. PLoS Pathog. 2020. PMID: 32598377 Free PMC article.
-
Comparison of Two Multilocus Sequence Typing Schemes for Mycoplasma bovis and Revision of the PubMLST Reference Method.J Clin Microbiol. 2020 May 26;58(6):e00283-20. doi: 10.1128/JCM.00283-20. Print 2020 May 26. J Clin Microbiol. 2020. PMID: 32295891 Free PMC article.
-
Extracellular vesicles of minimalistic Mollicutes as mediators of immune modulation and horizontal gene transfer.Commun Biol. 2025 Apr 29;8(1):674. doi: 10.1038/s42003-025-08099-4. Commun Biol. 2025. PMID: 40301684 Free PMC article.
-
De novo genome assembly resolving repetitive structures enables genomic analysis of 35 European Mycoplasmopsis bovis strains.BMC Genomics. 2023 Sep 16;24(1):548. doi: 10.1186/s12864-023-09618-5. BMC Genomics. 2023. PMID: 37715127 Free PMC article.
-
Improved transformation efficiency in Mycoplasma hominis enables disruption of the MIB-MIP system targeting human immunoglobulins.Microbiol Spectr. 2023 Sep 22;11(5):e0187323. doi: 10.1128/spectrum.01873-23. Online ahead of print. Microbiol Spectr. 2023. PMID: 37737635 Free PMC article.
References
-
- Aboklaish A. F., Dordet-Frisoni E., Citti C., Toleman M. A., Glass J. I., Spiller O. B. (2014). Random insertion and gene disruption via transposon mutagenesis of Ureaplasma parvum using a mini-transposon plasmid. Int. J. Med. Microbiol. 304 1218–1225. 10.1016/j.ijmm.2014.09.003 - DOI - PMC - PubMed

