Efficacy of Fecal Sampling as a Gut Proxy in the Study of Chicken Gut Microbiota
- PMID: 31572332
- PMCID: PMC6753641
- DOI: 10.3389/fmicb.2019.02126
Efficacy of Fecal Sampling as a Gut Proxy in the Study of Chicken Gut Microbiota
Abstract
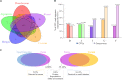
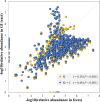
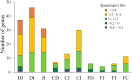
Despite the convenience and non-invasiveness of fecal sampling, the fecal microbiota does not fully represent that of the gastrointestinal (GI) tract, and the efficacy of fecal sampling to accurately represent the gut microbiota in birds is poorly understood. In this study, we aim to identify the efficacy of feces as a gut proxy in birds using chickens as a model. We collected 1,026 samples from 206 chickens, including duodenum, jejunum, ileum, cecum, and feces samples, for 16S rRNA amplicon sequencing analyses. In this study, the efficacy of feces as a gut proxy was partitioned to microbial community membership and community structure. Most taxa in the small intestine (84.11-87.28%) and ceca (99.39%) could be identified in feces. Microbial community membership was reflected with a gut anatomic feature, but community structure was not. Excluding shared microbes, the small intestine and ceca contributed 34.12 and 5.83% of the total fecal members, respectively. The composition of Firmicutes members in the small intestine and that of Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria members in the ceca could be well mirrored by the observations in fecal samples (ρ = 0.54-0.71 and 0.71-0.78, respectively, P < 0.001). However, there were few significant correlations for each genus between feces and each of the four gut segments, and these correlations were not high (ρ = -0.2-0.4, P < 0.05) for most genera. Our results suggest that fecal microbial community has a good potential to identify most taxa in the chicken gut and could moderately mirror the microbial structure in the intestine at the microbial population level with phylum specificity. However, it should be interpreted with caution by using feces as a proxy to study associations for microbial structure at individual microorganism level.
Keywords: chicken; feces; gut microbiota; proxy; spatial relationships.
Copyright © 2019 Yan, Sun, Zheng, Wen, Ji, Zhang, Chen, Hou and Yang.
Figures

Similar articles
-
Gastrointestinal Biogeography of Luminal Microbiota and Short-Chain Fatty Acids in Sika Deer (Cervus nippon).Appl Environ Microbiol. 2022 Sep 13;88(17):e0049922. doi: 10.1128/aem.00499-22. Epub 2022 Aug 11. Appl Environ Microbiol. 2022. PMID: 35950850 Free PMC article.
-
Spatial variations in the microbiota: comparative analysis of microbial composition and predicted functions across different intestinal segments and feces in donkeys.Front Microbiol. 2025 Jan 17;15:1494926. doi: 10.3389/fmicb.2024.1494926. eCollection 2024. Front Microbiol. 2025. PMID: 39895934 Free PMC article.
-
Microbial community mapping in intestinal tract of broiler chicken.Poult Sci. 2017 May 1;96(5):1387-1393. doi: 10.3382/ps/pew372. Poult Sci. 2017. PMID: 28339527
-
Microbial diversity and metabolic function in duodenum, jejunum and ileum of emu (Dromaius novaehollandiae).Sci Rep. 2023 Mar 18;13(1):4488. doi: 10.1038/s41598-023-31684-8. Sci Rep. 2023. PMID: 36934111 Free PMC article.
-
The temporal fluctuations and development of faecal microbiota in commercial layer flocks.Anim Nutr. 2023 Aug 9;15:197-209. doi: 10.1016/j.aninu.2023.07.006. eCollection 2023 Dec. Anim Nutr. 2023. PMID: 38023383 Free PMC article. Review.
Cited by
-
The regional diversity of gut microbiome along the GI tract of male C57BL/6 mice.BMC Microbiol. 2021 Feb 12;21(1):44. doi: 10.1186/s12866-021-02099-0. BMC Microbiol. 2021. PMID: 33579191 Free PMC article.
-
Toward Best Practice in Livestock Microbiota Research: A Comprehensive Comparison of Sample Storage and DNA Extraction Strategies.Front Microbiol. 2021 Feb 23;12:627539. doi: 10.3389/fmicb.2021.627539. eCollection 2021. Front Microbiol. 2021. PMID: 33708184 Free PMC article.
-
Taxonomy and Functional Diversity in the Fecal Microbiome of Beef Cattle Reared in Brazilian Traditional and Semi-Intensive Production Systems.Front Microbiol. 2021 Dec 8;12:768480. doi: 10.3389/fmicb.2021.768480. eCollection 2021. Front Microbiol. 2021. PMID: 34956130 Free PMC article.
-
The Effects of Vegetarian and Vegan Diets on Gut Microbiota.Front Nutr. 2019 Apr 17;6:47. doi: 10.3389/fnut.2019.00047. eCollection 2019. Front Nutr. 2019. PMID: 31058160 Free PMC article. Review.
-
Potential improvements of the cognition of piglets through a synbiotic supplementation from 1 to 28 days via the gut microbiota.Sci Rep. 2021 Dec 16;11(1):24113. doi: 10.1038/s41598-021-03565-5. Sci Rep. 2021. PMID: 34916559 Free PMC article.
References
-
- Alshamy Z., Richardson K. C., Hünigen H., Hafez H. M., Plendl J., Al Masri S. (2018). Comparison of the gastrointestinal tract of a dual-purpose to a broiler chicken line: a qualitative and quantitative macroscopic and microscopic study. PLoS One 13:e204921. 10.1371/journal.pone.0204921 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

