Life After Secretion- Yersinia enterocolitica Rapidly Toggles Effector Secretion and Can Resume Cell Division in Response to Changing External Conditions
- PMID: 31572334
- PMCID: PMC6753693
- DOI: 10.3389/fmicb.2019.02128
Life After Secretion- Yersinia enterocolitica Rapidly Toggles Effector Secretion and Can Resume Cell Division in Response to Changing External Conditions
Abstract
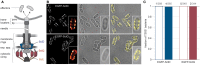
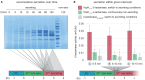
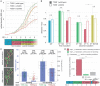
Many pathogenic bacteria use the type III secretion system (T3SS) injectisome to manipulate host cells by injecting virulence-promoting effector proteins into the host cytosol. The T3SS is activated upon host cell contact, and its activation is accompanied by an arrest of cell division; hence, many species maintain a T3SS-inactive sibling population to propagate efficiently within the host. The enteric pathogen Yersinia enterocolitica utilizes the T3SS to prevent phagocytosis and inhibit inflammatory responses. Unlike other species, almost all Y. enterocolitica are T3SS-positive at 37°C, which raises the question, how these bacteria are able to propagate within the host, that is, when and how they stop secretion and restart cell division after a burst of secretion. Using a fast and quantitative in vitro secretion assay, we have examined the initiation and termination of type III secretion. We found that effector secretion begins immediately once the activating signal is present, and instantly stops when this signal is removed. Following effector secretion, the bacteria resume division within minutes after being introduced to a non-secreting environment, and the same bacteria are able to re-initiate effector secretion at later time points. Our results indicate that Y. enterocolitica use their type III secretion system to promote their individual survival when necessary, and are able to quickly switch their behavior toward replication afterwards, possibly gaining an advantage during infection.
Keywords: Yersinia enterocolitica; bacterial protein secretion; enteropathogens; host-pathogen interaction; protein translocation; regulation of virulence mechanisms.
Copyright © 2019 Milne-Davies, Helbig, Wimmi, Cheng, Paczia and Diepold.
Figures

References
LinkOut - more resources
Full Text Sources

