A Single-Stranded Oligonucleotide Inhibits Toll-Like Receptor 3 Activation and Reduces Influenza A (H1N1) Infection
- PMID: 31572376
- PMCID: PMC6751283
- DOI: 10.3389/fimmu.2019.02161
A Single-Stranded Oligonucleotide Inhibits Toll-Like Receptor 3 Activation and Reduces Influenza A (H1N1) Infection
Abstract
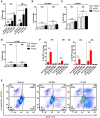
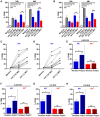
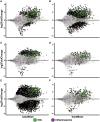
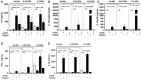
The initiation of an immune response is dependent on the activation and maturation of dendritic cells after sensing pathogen associated molecular patterns by pattern recognition receptors. However, the response needs to be balanced as excessive pro-inflammatory cytokine production in response to viral or stress-induced pattern recognition receptor signaling has been associated with severe influenza A virus (IAV) infection. Here, we use an inhibitor of Toll-like receptor (TLR)3, a single-stranded oligonucleotide (ssON) with the capacity to inhibit certain endocytic routes, or a TLR3 agonist (synthetic double-stranded RNA PolyI:C), to evaluate modulation of innate responses during H1N1 IAV infection. Since IAV utilizes cellular endocytic machinery for viral entry, we also assessed ssON's capacity to affect IAV infection. We first show that IAV infected human monocyte-derived dendritic cells (MoDC) were unable to up-regulate the co-stimulatory molecules CD80 and CD86 required for T cell activation. Exogenous TLR3 stimulation did not overcome the IAV-mediated inhibition of co-stimulatory molecule expression in MoDC. However, TLR3 stimulation using PolyI:C led to an augmented pro-inflammatory cytokine response. We reveal that ssON effectively inhibited PolyI:C-mediated pro-inflammatory cytokine production in MoDC, notably, ssON treatment maintained an interferon response induced by IAV infection. Accordingly, RNAseq analyses revealed robust up-regulation of interferon-stimulated genes in IAV cultures treated with ssON. We next measured reduced IAV production in MoDC treated with ssON and found a length requirement for its anti-viral activity, which overlapped with its capacity to inhibit uptake of PolyI:C. Hence, in cases wherein an overreacting TLR3 activation contributes to IAV pathogenesis, ssON can reduce this signaling pathway. Furthermore, concomitant treatment with ssON and IAV infection in mice resulted in maintained weight and reduced viral load in the lungs. Therefore, extracellular ssON provides a mechanism for immune regulation of TLR3-mediated responses and suppression of IAV infection in vitro and in vivo in mice.
Keywords: TLR3; clathrin-mediated endocytosis; co-stimulatory molecules; cytokines; human monocyte-derived dendritic cells (MoDC); influenza A; mice; single-stranded oligonucleotides.
Copyright © 2019 Poux, Dondalska, Bergenstråhle, Pålsson, Contreras, Arasa, Järver, Albert, Busse, LeGrand, Lundeberg, Tregoning and Spetz.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

