The regulation of chromosome segregation via centromere loops
- PMID: 31573359
- PMCID: PMC6856439
- DOI: 10.1080/10409238.2019.1670130
The regulation of chromosome segregation via centromere loops
Abstract
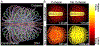
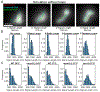
Biophysical studies of the yeast centromere have shown that the organization of the centromeric chromatin plays a crucial role in maintaining proper tension between sister kinetochores during mitosis. While centromeric chromatin has traditionally been considered a simple spring, recent work reveals the centromere as a multifaceted, tunable shock absorber. Centromeres can differ from other regions of the genome in their heterochromatin state, supercoiling state, and enrichment of structural maintenance of chromosomes (SMC) protein complexes. Each of these differences can be utilized to alter the effective stiffness of centromeric chromatin. In budding yeast, the SMC protein complexes condensin and cohesin stiffen chromatin by forming and cross-linking chromatin loops, respectively, into a fibrous structure resembling a bottlebrush. The high density of the loops compacts chromatin while spatially isolating the tension from spindle pulling forces to a subset of the chromatin. Paradoxically, the molecular crowding of chromatin via cohesin and condensin also causes an outward/poleward force. The structure allows the centromere to act as a shock absorber that buffers the variable forces generated by dynamic spindle microtubules. Based on the distribution of SMCs from bacteria to human and the conserved distance between sister kinetochores in a wide variety of organisms (0.4 to 1 micron), we propose that the bottlebrush mechanism is the foundational principle for centromere function in eukaryotes.
Keywords: Centromere; DNA loops; chromosome segregation; cohesin; condensin; kinetochore; mitosis; pericentromere.
Figures
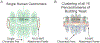
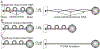
Similar articles
-
Shaping centromeres to resist mitotic spindle forces.J Cell Sci. 2022 Feb 15;135(4):jcs259532. doi: 10.1242/jcs.259532. Epub 2022 Feb 18. J Cell Sci. 2022. PMID: 35179192 Free PMC article.
-
Individual pericentromeres display coordinated motion and stretching in the yeast spindle.J Cell Biol. 2013 Nov 11;203(3):407-16. doi: 10.1083/jcb.201307104. Epub 2013 Nov 4. J Cell Biol. 2013. PMID: 24189271 Free PMC article.
-
ChromoShake: a chromosome dynamics simulator reveals that chromatin loops stiffen centromeric chromatin.Mol Biol Cell. 2016 Jan 1;27(1):153-66. doi: 10.1091/mbc.E15-08-0575. Epub 2015 Nov 4. Mol Biol Cell. 2016. PMID: 26538024 Free PMC article.
-
Condensin in Chromatid Cohesion and Segregation.Cytogenet Genome Res. 2015;147(4):212-6. doi: 10.1159/000444868. Epub 2016 Mar 22. Cytogenet Genome Res. 2015. PMID: 26998746 Review.
-
Centromeric heterochromatin: the primordial segregation machine.Annu Rev Genet. 2014;48:457-84. doi: 10.1146/annurev-genet-120213-092033. Epub 2014 Sep 18. Annu Rev Genet. 2014. PMID: 25251850 Free PMC article. Review.
Cited by
-
It's all in the numbers: Cohesin stoichiometry.Front Mol Biosci. 2022 Oct 18;9:1010894. doi: 10.3389/fmolb.2022.1010894. eCollection 2022. Front Mol Biosci. 2022. PMID: 36330215 Free PMC article. Review.
-
R-loops at centromeric chromatin contribute to defects in kinetochore integrity and chromosomal instability in budding yeast.Mol Biol Cell. 2021 Jan 1;32(1):74-89. doi: 10.1091/mbc.E20-06-0379. Epub 2020 Nov 4. Mol Biol Cell. 2021. PMID: 33147102 Free PMC article.
-
Polymer Modeling Reveals Interplay between Physical Properties of Chromosomal DNA and the Size and Distribution of Condensin-Based Chromatin Loops.Genes (Basel). 2023 Dec 9;14(12):2193. doi: 10.3390/genes14122193. Genes (Basel). 2023. PMID: 38137015 Free PMC article.
-
Behavior of dicentric chromosomes in budding yeast.PLoS Genet. 2021 Mar 18;17(3):e1009442. doi: 10.1371/journal.pgen.1009442. eCollection 2021 Mar. PLoS Genet. 2021. PMID: 33735169 Free PMC article.
-
Independence of centromeric and pericentromeric chromatin stability on CCAN components.Mol Biol Cell. 2025 Apr 1;36(4):ar41. doi: 10.1091/mbc.E24-02-0066. Epub 2025 Feb 12. Mol Biol Cell. 2025. PMID: 39937678 Free PMC article.
References
-
- Flemming W (1882). Zellsubstanz, Kern und Zelltheilung (Cell substance, nucleus and cell division). Leipzig, Vogel, 1882
-
- Clarke L, Carbon J. (1980). Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature, 287, 504–9 - PubMed
-
- Fitzgerald-Hayes M, Clarke L, Carbon J. (1982). Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell, 29, 235–44 - PubMed
-
- Blat Y, Kleckner N. (1999). Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell, 98, 249–59 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
