H3K9me2 orchestrates inheritance of spatial positioning of peripheral heterochromatin through mitosis
- PMID: 31573510
- PMCID: PMC6795522
- DOI: 10.7554/eLife.49278
H3K9me2 orchestrates inheritance of spatial positioning of peripheral heterochromatin through mitosis
Abstract
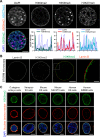
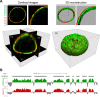
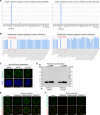
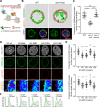
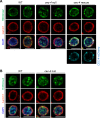
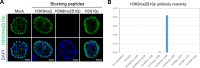
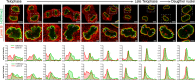
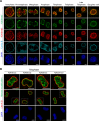
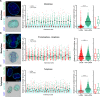
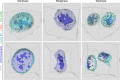
Cell-type-specific 3D organization of the genome is unrecognizable during mitosis. It remains unclear how essential positional information is transmitted through cell division such that a daughter cell recapitulates the spatial genome organization of the parent. Lamina-associated domains (LADs) are regions of repressive heterochromatin positioned at the nuclear periphery that vary by cell type and contribute to cell-specific gene expression and identity. Here we show that histone 3 lysine 9 dimethylation (H3K9me2) is an evolutionarily conserved, specific mark of nuclear peripheral heterochromatin and that it is retained through mitosis. During mitosis, phosphorylation of histone 3 serine 10 temporarily shields the H3K9me2 mark allowing for dissociation of chromatin from the nuclear lamina. Using high-resolution 3D immuno-oligoFISH, we demonstrate that H3K9me2-enriched genomic regions, which are positioned at the nuclear lamina in interphase cells prior to mitosis, re-associate with the forming nuclear lamina before mitotic exit. The H3K9me2 modification of peripheral heterochromatin ensures that positional information is safeguarded through cell division such that individual LADs are re-established at the nuclear periphery in daughter nuclei. Thus, H3K9me2 acts as a 3D architectural mitotic guidepost. Our data establish a mechanism for epigenetic memory and inheritance of spatial organization of the genome.
Keywords: C. elegans; H3K9me2; cell biology; chromosomes; gene expression; genome organization; mitosis; mouse; nuclear architecture; nuclear lamina; peripheral heterochromatin.
© 2019, Poleshko et al.
Conflict of interest statement
AP, CS, SN, PS, KW, JM, ML, EJ, RJ, JE No competing interests declared
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

