Combinatorial mutagenesis of rapidly evolving residues yields super-restrictor antiviral proteins
- PMID: 31574080
- PMCID: PMC6772013
- DOI: 10.1371/journal.pbio.3000181
Combinatorial mutagenesis of rapidly evolving residues yields super-restrictor antiviral proteins
Abstract
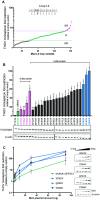
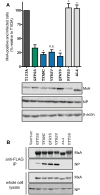
Antagonistic interactions drive host-virus evolutionary arms races, which often manifest as recurrent amino acid changes (i.e., positive selection) at their protein-protein interaction interfaces. Here, we investigated whether combinatorial mutagenesis of positions under positive selection in a host antiviral protein could enhance its restrictive properties. We tested approximately 700 variants of human MxA, generated by combinatorial mutagenesis, for their ability to restrict Thogotovirus (THOV). We identified MxA super-restrictors with increased binding to the THOV nucleoprotein (NP) target protein and 10-fold higher anti-THOV restriction relative to wild-type human MxA, the most potent naturally occurring anti-THOV restrictor identified. Our findings reveal a means to elicit super-restrictor antiviral proteins by leveraging signatures of positive selection. Although some MxA super-restrictors of THOV were impaired in their restriction of H5N1 influenza A virus (IAV), other super-restrictor variants increased THOV restriction without impairment of IAV restriction. Thus, broadly acting antiviral proteins such as MxA mitigate breadth-versus-specificity trade-offs that could otherwise constrain their adaptive landscape.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Tick-transmitted thogotovirus gains high virulence by a single MxA escape mutation in the viral nucleoprotein.PLoS Pathog. 2020 Nov 16;16(11):e1009038. doi: 10.1371/journal.ppat.1009038. eCollection 2020 Nov. PLoS Pathog. 2020. PMID: 33196685 Free PMC article.
-
Heterozygous and generalist MxA super-restrictors overcome breadth-specificity trade-offs in antiviral restriction.Sci Adv. 2025 May 2;11(18):eadu0062. doi: 10.1126/sciadv.adu0062. Epub 2025 May 2. Sci Adv. 2025. PMID: 40315333 Free PMC article.
-
Heterozygous and generalist MxA super-restrictors overcome breadth-specificity tradeoffs in antiviral restriction.bioRxiv [Preprint]. 2024 Oct 10:2024.10.10.617484. doi: 10.1101/2024.10.10.617484. bioRxiv. 2024. Update in: Sci Adv. 2025 May 2;11(18):eadu0062. doi: 10.1126/sciadv.adu0062. PMID: 39416221 Free PMC article. Updated. Preprint.
-
An evolutionary perspective on the broad antiviral specificity of MxA.Curr Opin Microbiol. 2013 Aug;16(4):493-9. doi: 10.1016/j.mib.2013.04.005. Epub 2013 May 28. Curr Opin Microbiol. 2013. PMID: 23725670 Free PMC article. Review.
-
Human MxA protein: an interferon-induced dynamin-like GTPase with broad antiviral activity.J Interferon Cytokine Res. 2011 Jan;31(1):79-87. doi: 10.1089/jir.2010.0076. Epub 2010 Dec 19. J Interferon Cytokine Res. 2011. PMID: 21166595 Review.
Cited by
-
A Rapidly Evolving Polybasic Motif Modulates Bacterial Detection by Guanylate Binding Proteins.mBio. 2020 May 19;11(3):e00340-20. doi: 10.1128/mBio.00340-20. mBio. 2020. PMID: 32430466 Free PMC article.
-
Tick-transmitted thogotovirus gains high virulence by a single MxA escape mutation in the viral nucleoprotein.PLoS Pathog. 2020 Nov 16;16(11):e1009038. doi: 10.1371/journal.ppat.1009038. eCollection 2020 Nov. PLoS Pathog. 2020. PMID: 33196685 Free PMC article.
-
Systematic genetic characterization of the human PKR kinase domain highlights its functional malleability to escape a poxvirus substrate mimic.bioRxiv [Preprint]. 2024 Sep 22:2024.05.29.596416. doi: 10.1101/2024.05.29.596416. bioRxiv. 2024. Update in: Elife. 2024 Nov 12;13:RP99575. doi: 10.7554/eLife.99575. PMID: 38903081 Free PMC article. Updated. Preprint.
-
Conservation lost: host-pathogen battles drive diversification and expansion of gene families.FEBS J. 2021 Sep;288(18):5289-5299. doi: 10.1111/febs.15627. Epub 2020 Dec 2. FEBS J. 2021. PMID: 33190369 Free PMC article. Review.
-
RTP4 Is a Potent IFN-Inducible Anti-flavivirus Effector Engaged in a Host-Virus Arms Race in Bats and Other Mammals.Cell Host Microbe. 2020 Nov 11;28(5):712-723.e9. doi: 10.1016/j.chom.2020.09.014. Epub 2020 Oct 27. Cell Host Microbe. 2020. PMID: 33113352 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

