A Prediction Model to Help with Oncologic Mediastinal Evaluation for Radiation: HOMER
- PMID: 31574238
- PMCID: PMC6961739
- DOI: 10.1164/rccm.201904-0831OC
A Prediction Model to Help with Oncologic Mediastinal Evaluation for Radiation: HOMER
Abstract
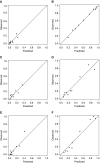
Rationale: When stereotactic ablative radiotherapy is an option for patients with non-small cell lung cancer (NSCLC), distinguishing between N0, N1, and N2 or N3 (N2|3) disease is important.Objectives: To develop a prediction model for estimating the probability of N0, N1, and N2|3 disease.Methods: Consecutive patients with clinical-radiographic stage T1 to T3, N0 to N3, and M0 NSCLC who underwent endobronchial ultrasound-guided staging from a single center were included. Multivariate ordinal logistic regression analysis was used to predict the presence of N0, N1, or N2|3 disease. Temporal validation used consecutive patients from 3 years later at the same center. External validation used three other hospitals.Measurements and Main Results: In the model development cohort (n = 633), younger age, central location, adenocarcinoma, and higher positron emission tomography-computed tomography nodal stage were associated with a higher probability of having advanced nodal disease. Areas under the receiver operating characteristic curve (AUCs) were 0.84 and 0.86 for predicting N1 or higher (vs. N0) disease and N2|3 (vs. N0 or N1) disease, respectively. Model fit was acceptable (Hosmer-Lemeshow, P = 0.960; Brier score, 0.36). In the temporal validation cohort (n = 473), AUCs were 0.86 and 0.88. Model fit was acceptable (Hosmer-Lemeshow, P = 0.172; Brier score, 0.30). In the external validation cohort (n = 722), AUCs were 0.86 and 0.88 but required calibration (Hosmer-Lemeshow, P < 0.001; Brier score, 0.38). Calibration using the general calibration method resulted in acceptable model fit (Hosmer-Lemeshow, P = 0.094; Brier score, 0.34).Conclusions: This prediction model can estimate the probability of N0, N1, and N2|3 disease in patients with NSCLC. The model has the potential to facilitate decision-making in patients with NSCLC when stereotactic ablative radiotherapy is an option.
Keywords: endobronchial ultrasound; lung cancer; lung cancer staging; mediastinal adenopathy.
Figures
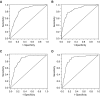
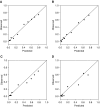
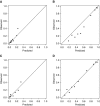
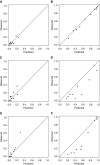
Comment in
-
Hitting a HOMER: Epidemiology to the Bedside when Evaluating for Stereotactic Ablative Radiotherapy.Am J Respir Crit Care Med. 2020 Jan 15;201(2):136-138. doi: 10.1164/rccm.201910-1933ED. Am J Respir Crit Care Med. 2020. PMID: 31658428 Free PMC article. No abstract available.
References
-
- Silvestri GA, Gonzalez AV, Jantz MA, Margolis ML, Gould MK, Tanoue LT, et al. Methods for staging non-small cell lung cancer: diagnosis and management of lung cancer, 3rd ed. American College of Chest Physicians evidence-based clinical practice guidelines. Chest. 2013;143:e211S–e250S. - PubMed
-
- National Comprehensive Cancer Network. Non-small cell lung cancer. Version 2.2019; 2018 [accessed 2019 Dec 13]. Available from: https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf.
-
- Reck M, Rabe KF. Precision diagnosis and treatment for advanced non–small-cell lung cancer. N Engl J Med. 2017;377:849–861. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

