Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition
- PMID: 31575017
- PMCID: PMC6829226
- DOI: 10.3390/cells8101178
Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition
Abstract
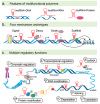
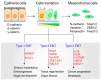
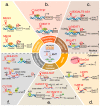
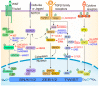
Long non-coding RNAs (lncRNAs) are versatile regulators of gene expression and play crucial roles in diverse biological processes. Epithelial-mesenchymal transition (EMT) is a cellular program that drives plasticity during embryogenesis, wound healing, and malignant progression. Increasing evidence shows that lncRNAs orchestrate multiple cellular processes by modulating EMT in diverse cell types. Dysregulated lncRNAs that can impact epithelial plasticity by affecting different EMT markers and target genes have been identified. However, our understanding of the landscape of lncRNAs important in EMT is far from complete. Here, we summarize recent findings on the mechanisms and roles of lncRNAs in EMT and elaborate on how lncRNAs can modulate EMT by interacting with RNA, DNA, or proteins in epigenetic, transcriptional, and post-transcriptional regulation. This review also highlights significant EMT pathways that may be altered by diverse lncRNAs, thereby suggesting their therapeutic potential.
Keywords: cancer; epithelial-mesenchymal transition (EMT); long non-coding RNA; signaling pathways.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

