The TCP4 Transcription Factor Directly Activates TRICHOMELESS1 and 2 and Suppresses Trichome Initiation
- PMID: 31575625
- PMCID: PMC6878003
- DOI: 10.1104/pp.19.00197
The TCP4 Transcription Factor Directly Activates TRICHOMELESS1 and 2 and Suppresses Trichome Initiation
Abstract
Trichomes are the first line of defense on the outer surface of plants against biotic and abiotic stresses. Because trichomes on leaf surfaces originate from the common epidermal progenitor cells that also give rise to pavement cells and stomata, their density and distribution are under strict genetic control. Regulators of trichome initiation have been identified and incorporated into a biochemical pathway wherein an initiator complex promotes trichome fate in an epidermal progenitor cell, while an inhibitor complex suppresses it in the neighboring cells. However, it is unclear how these regulator proteins, especially the negative regulators, are induced by upstream transcription factors and integrated with leaf morphogenesis. Here, we show that the Arabidopsis (Arabidopsis thaliana) class II TCP proteins activate TRICHOMELESS1 (TCL1) and TCL2, the two established negative regulators of trichome initiation, and reduce trichome density on leaves. Loss-of-function of these TCP proteins increased trichome density whereas TCP4 gain-of-function reduced trichome number. TCP4 binds to the upstream regulatory elements of both TCL1 and TCL 2 and directly promotes their transcription. Further, the TCP-induced trichome suppression is independent of the SQUAMOSA PROMOTER BINDING PROTEIN LIKE family of transcription factors, proteins that also reduce trichome density at later stages of plant development. Our work demonstrates that the class II TCP proteins couple leaf morphogenesis with epidermal cell fate determination.
© 2019 American Society of Plant Biologists. All Rights Reserved.
Figures


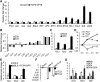


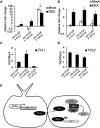
Comment in
-
A Novel Upstream Regulator of Trichome Development Inhibitors.Plant Physiol. 2019 Dec;181(4):1398-1400. doi: 10.1104/pp.19.01269. Plant Physiol. 2019. PMID: 31767791 Free PMC article. No abstract available.
References
-
- Aggarwal P, Challa KR, Rath M, Sunkara P, Nath U (2018) Generation of inducible transgenic lines of Arabidopsis transcription factors regulated by microRNAs. Methods Mol Biol 1830: 61–79 - PubMed
-
- Balkunde R, Pesch M, Hülskamp M (2010) Trichome patterning in Arabidopsis thaliana from genetic to molecular models. Curr Top Dev Biol 91: 299–321 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

