Molecular Inhibitor of QSOX1 Suppresses Tumor Growth In Vivo
- PMID: 31575656
- PMCID: PMC6946859
- DOI: 10.1158/1535-7163.MCT-19-0233
Molecular Inhibitor of QSOX1 Suppresses Tumor Growth In Vivo
Abstract
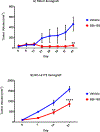
Quiescin sulfhydryl oxidase 1 (QSOX1) is an enzyme overexpressed by many different tumor types. QSOX1 catalyzes the formation of disulfide bonds in proteins. Because short hairpin knockdowns (KD) of QSOX1 have been shown to suppress tumor growth and invasion in vitro and in vivo, we hypothesized that chemical compounds inhibiting QSOX1 enzymatic activity would also suppress tumor growth, invasion, and metastasis. High throughput screening using a QSOX1-based enzymatic assay revealed multiple potential QSOX1 inhibitors. One of the inhibitors, known as "SBI-183," suppresses tumor cell growth in a Matrigel-based spheroid assay and inhibits invasion in a modified Boyden chamber, but does not affect viability of nonmalignant cells. Oral administration of SBI-183 inhibits tumor growth in 2 independent human xenograft mouse models of renal cell carcinoma. We conclude that SBI-183 warrants further exploration as a useful tool for understanding QSOX1 biology and as a potential novel anticancer agent in tumors that overexpress QSOX1.
©2019 American Association for Cancer Research.
Conflict of interest statement
Figures




References
-
- T. E Gilles C, Newgreen DF, Sato H, “Matrix Metalloproteases and Epithelial-to-Mesenchymal Transition: Implications for Carcinoma Metastasis,” in Madame Curie Bioscience Database [Internet]., Austin (TX): Landes Bioscience.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

