2'-C-methylated nucleotides terminate virus RNA synthesis by preventing active site closure of the viral RNA-dependent RNA polymerase
- PMID: 31575662
- PMCID: PMC6851289
- DOI: 10.1074/jbc.RA119.010214
2'-C-methylated nucleotides terminate virus RNA synthesis by preventing active site closure of the viral RNA-dependent RNA polymerase
Abstract
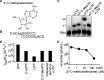
The 2'-C-methyl ribonucleosides are nucleoside analogs representing an important class of antiviral agents, especially against positive-strand RNA viruses. Their value is highlighted by the highly successful anti-hepatitis C drug sofosbuvir. When appropriately phosphorylated, these nucleotides are successfully incorporated into RNA by the virally encoded RNA-dependent RNA polymerase (RdRp). This activity prevents further RNA extension, but the mechanism is poorly characterized. Previously, we had identified NMR signatures characteristic of formation of RdRp-RNA binary and RdRp-RNA-NTP ternary complexes for the poliovirus RdRp, including an open-to-closed conformational change necessary to prepare the active site for catalysis of phosphoryl transfer. Here we used these observations as a framework for interpreting the effects of 2'-C-methyl adenosine analogs on RNA chain extension in solution-state NMR spectroscopy experiments, enabling us to gain additional mechanistic insights into 2'-C-methyl ribonucleoside-mediated RNA chain termination. Contrary to what has been proposed previously, poliovirus RdRp that was bound to RNA with an incorporated 2'-C-methyl nucleotide could still bind to the next incoming NTP. Our results also indicated that incorporation of the 2'-C-methyl nucleotide does not disrupt RdRp-RNA interactions and does not prevent translocation. Instead, incorporation of the 2'-C-methyl nucleotide blocked closure of the RdRp active site upon binding of the next correct incoming NTP, which prevented further nucleotide addition. We propose that other nucleotide analogs that act as nonobligate chain terminators may operate through a similar mechanism.
Keywords: 2′-C-methyl ribonucleoside; NMR; RNA polymerase; antiviral agent; hepatitis C virus (HCV); nucleoside/nucleotide analogue; plus-stranded RNA virus; poliovirus; positive-strand RNA virus; viral polymerase.
© 2019 Boehr et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health
Figures
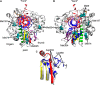






References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

