Chemoperception of Specific Amino Acids Controls Phytopathogenicity in Pseudomonas syringae pv. tomato
- PMID: 31575767
- PMCID: PMC6775455
- DOI: 10.1128/mBio.01868-19
Chemoperception of Specific Amino Acids Controls Phytopathogenicity in Pseudomonas syringae pv. tomato
Abstract
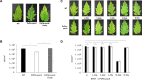
Chemotaxis has been associated with the pathogenicity of bacteria in plants and was found to facilitate bacterial entry through stomata and wounds. However, knowledge regarding the plant signals involved in this process is scarce. We have addressed this issue using Pseudomonas syringae pv. tomato, which is a foliar pathogen that causes bacterial speck in tomato. We show that the chemoreceptor P. syringae pv. tomato PscA (PsPto-PscA) recognizes specifically and with high affinity l-Asp, l-Glu, and d-Asp. The mutation of the chemoreceptor gene largely reduced chemotaxis to these ligands but also altered cyclic di-GMP (c-di-GMP) levels, biofilm formation, and motility, pointing to cross talk between different chemosensory pathways. Furthermore, the PsPto-PscA mutant strain showed reduced virulence in tomato. Asp and Glu are the most abundant amino acids in plants and in particular in tomato apoplasts, and we hypothesize that this receptor may have evolved to specifically recognize these compounds to facilitate bacterial entry into the plant. Infection assays with the wild-type strain showed that the presence of saturating concentrations of d-Asp also reduced bacterial virulence.IMPORTANCE There is substantive evidence that chemotaxis is a key requisite for efficient pathogenesis in plant pathogens. However, information regarding particular bacterial chemoreceptors and the specific plant signal that they sense is scarce. Our work shows that the phytopathogenic bacterium Pseudomonas syringae pv. tomato mediates not only chemotaxis but also the control of pathogenicity through the perception of the plant abundant amino acids Asp and Glu. We describe the specificity of the perception of l- and d-Asp and l-Glu by the PsPto-PscA chemoreceptor and the involvement of this perception in the regulation of pathogenicity-related traits. Moreover, a saturating concentration of d-Asp reduces bacterial virulence, and we therefore propose that ligand-mediated interference of key chemoreceptors may be an alternative strategy to control virulence.
Keywords: Pseudomonas syringae; chemoreceptors; virulence.
Copyright © 2019 Cerna-Vargas et al.
Figures



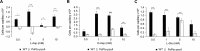




Similar articles
-
Pseudomonas syringae pv. tomato infection of tomato plants is mediated by GABA and l-Pro chemoperception.Mol Plant Pathol. 2022 Oct;23(10):1433-1445. doi: 10.1111/mpp.13238. Epub 2022 Jun 10. Mol Plant Pathol. 2022. PMID: 35689388 Free PMC article.
-
Chemosensory systems interact to shape relevant traits for bacterial plant pathogenesis.mBio. 2024 Jul 17;15(7):e0087124. doi: 10.1128/mbio.00871-24. Epub 2024 Jun 20. mBio. 2024. PMID: 38899869 Free PMC article.
-
Blue-light perception by epiphytic Pseudomonas syringae drives chemoreceptor expression, enabling efficient plant infection.Mol Plant Pathol. 2020 Dec;21(12):1606-1619. doi: 10.1111/mpp.13001. Epub 2020 Oct 7. Mol Plant Pathol. 2020. PMID: 33029921 Free PMC article.
-
Pseudomonas syringae type III secretion system effectors: repertoires in search of functions.Curr Opin Microbiol. 2009 Feb;12(1):53-60. doi: 10.1016/j.mib.2008.12.003. Epub 2009 Jan 23. Curr Opin Microbiol. 2009. PMID: 19168384 Review.
-
Defining essential processes in plant pathogenesis with Pseudomonas syringae pv. tomato DC3000 disarmed polymutants and a subset of key type III effectors.Mol Plant Pathol. 2018 Jul;19(7):1779-1794. doi: 10.1111/mpp.12655. Epub 2018 Feb 1. Mol Plant Pathol. 2018. PMID: 29277959 Free PMC article. Review.
Cited by
-
A bacterial chemoreceptor that mediates chemotaxis to two different plant hormones.Environ Microbiol. 2022 Aug;24(8):3580-3597. doi: 10.1111/1462-2920.15920. Epub 2022 Feb 1. Environ Microbiol. 2022. PMID: 35088505 Free PMC article.
-
Requirement of γ-Aminobutyric Acid Chemotaxis for Virulence of Pseudomonas syringae pv. tabaci 6605.Microbes Environ. 2020;35(4):ME20114. doi: 10.1264/jsme2.ME20114. Microbes Environ. 2020. PMID: 33162464 Free PMC article.
-
The Repertoire of Solute-Binding Proteins of Model Bacteria Reveals Large Differences in Number, Type, and Ligand Range.Microbiol Spectr. 2022 Oct 26;10(5):e0205422. doi: 10.1128/spectrum.02054-22. Epub 2022 Sep 19. Microbiol Spectr. 2022. PMID: 36121253 Free PMC article.
-
Biofilm Dispersal in Bacillus velezensis FZB42 Is Regulated by the Second Messenger c-di-GMP.Microorganisms. 2025 Apr 13;13(4):896. doi: 10.3390/microorganisms13040896. Microorganisms. 2025. PMID: 40284732 Free PMC article.
-
Pectobacterium brasiliense 1692 Chemotactic Responses and the Role of Methyl-Accepting Chemotactic Proteins in Ecological Fitness.Front Plant Sci. 2021 Apr 22;12:650894. doi: 10.3389/fpls.2021.650894. eCollection 2021. Front Plant Sci. 2021. PMID: 33968106 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
