Glycan-mediated enhancement of reovirus receptor binding
- PMID: 31575869
- PMCID: PMC6773860
- DOI: 10.1038/s41467-019-12411-2
Glycan-mediated enhancement of reovirus receptor binding
Abstract
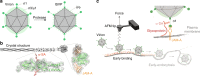
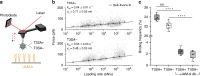
Viral infection is an intricate process that requires the concerted action of both viral and host cell components. Entry of viruses into cells is initiated by interactions between viral proteins and their cell surface receptors. Despite recent progress, the molecular mechanisms underlying the multistep reovirus entry process are poorly understood. Using atomic force microscopy, we investigated how the reovirus σ1 attachment protein binds to both α-linked sialic acid (α-SA) and JAM-A cell-surface receptors. We discovered that initial σ1 binding to α-SA favors a strong multivalent anchorage to JAM-A. The enhanced JAM-A binding by virions following α-SA engagement is comparable to JAM-A binding by infectious subvirion particles (ISVPs) in the absence of α-SA. Since ISVPs have an extended σ1 conformer, this finding suggests that α-SA binding triggers a conformational change in σ1. These results provide new insights into the function of viral attachment proteins in the initiation of infection and open new avenues for the use of reoviruses as oncolytic agents.
Conflict of interest statement
M.K. and D.A. have applied for a patent for the use of sialylated glycans in combination with reoviruses (EP19152640.9). The remaining authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

