Activity-Based Sensing of S-Depalmitoylases: Chemical Technologies and Biological Discovery
- PMID: 31577124
- PMCID: PMC7201403
- DOI: 10.1021/acs.accounts.9b00354
Activity-Based Sensing of S-Depalmitoylases: Chemical Technologies and Biological Discovery
Abstract
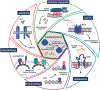
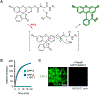
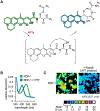
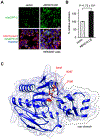
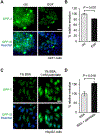
While lipids were first appreciated as a critical hydrophobic barrier, our understanding of their roles at the cellular and organismal levels continues to grow. Not only are they important independent operators, providing a platform for both static and dynamic organization and communication within the cell, they also exert significant effects via the chemical modification of proteins. Addition of a lipid post-translational modification (PTM) alters protein hydrophobicity and behavior, with distinct consequences for subcellular trafficking, localization, intra- and intermolecular interactions, and stability. One of the most abundant and widespread protein lipidation events is S-acylation, installation of a long-chain lipid to the thiol of a cysteine side chain through a thioester linkage. S-Acylation is often referred to as S-palmitoylation, due to the prevalence of palmitate as the lipid modification. Unlike many lipid PTMs, S-acylation is enzymatically reversible, enabling the cell to tune proteome-wide properties through dynamic alterations in protein lipidation status. While much has been uncovered about the molecular effects of S-acylation and its implications for physiology, current biochemical and chemical methods only assess substrate lipidation levels or steady-state levels of enzyme activity. Yet, the writer protein acyl transferases (PATs) and eraser acyl protein thioesterases (APTs) are dynamically active, responsible for sometimes-rapid changes in S-palmitoylation status of target proteins. Thus, to understand the full scope, significance, and subtlety of S-deacylation and its regulation in the cell, it is necessary to observe the timing and cellular geography of regulatory enzyme activities. In this Account, we review the chemical tools developed by our group to selectively visualize and perturb the activity of APTs in live cells, highlighting the biological insights gained from their application. To visualize APT activity, we masked fluorogenic molecules with thioacylated, peptide-based APT substrate mimetics; APT activity and thus thiol deprotection releases a fluorescent product in the turn-on depalmitoylation probes (DPPs), while in ratiometric depalmitoylation probes (RDPs) the emission of the parent fluorophore is altered. Application of these probes in live cells reveals that APT activity is sensitive to cell signaling events and metabolic disturbances. Additionally, as indicated above, the location of regulatory enzymes is critical in lipid signaling, and one organelle of particular interest, due to its role in maintaining cellular homeostasis and its legion of lipidated proteins, is the mitochondria. Therefore, we developed a class of spatially constrained mitoDPPs to visualize mitochondrial APT activity as well as a selective inhibitor of mitochondrial deacylation activity, mitoFP. With these tools, we identify two mitochondrial S-depalmitoylases and connect mitochondrial S-depalmitoylation to redox buffering capacity. Moreover, some of the changes in activity observed are specific to the mitochondria, confirming spatial as well as temporal regulation of eraser protein activity. Overall, this chemical toolkit for S-depalmitoylase activity, imaging reagents and a targeted inhibitor, will continue to illuminate the regulatory mechanisms and roles of S-depalmitoylation within the complex homeostatic networks of the cell.
Conflict of interest statement
Competing interests
B.C.D. and R.S.K. have a patent on the DPPs.
Figures
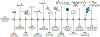

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

