Determinants of Active-Site Inhibitor Interaction with HIV-1 RNase H
- PMID: 31577424
- PMCID: PMC6842066
- DOI: 10.1021/acsinfecdis.9b00300
Determinants of Active-Site Inhibitor Interaction with HIV-1 RNase H
Abstract
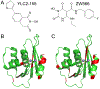
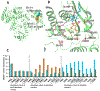
The ribonuclease H (RNH) activity of HIV-1 reverse transcriptase (RT) is essential for viral replication and can be a target for drug development. Yet, no RNH inhibitor to date has substantial antiviral activity to allow advancement into clinical development. Herein, we describe our characterization of the detailed binding mechanisms of RNH active-site inhibitors, YLC2-155 and ZW566, that bind to the RNH domain through divalent metal ions, using NMR, molecular docking, and quantum mechanical calculations. In the presence of Mg2+, NMR spectra of RNH exhibited split (two) resonances for some residues upon inhibitor binding, suggesting two binding modes, an observation consistent with the docking results. The relative populations of the two binding conformers were independent of inhibitor or Mg2+ concentration, with one conformation consistently more favored. In our docking study, one distinctive pose of ZW566 showed more interactions with surrounding residues of RNH compared to the analogous binding pose of YLC2-155. Inhibitor titration experiments revealed a lower dissociation constant for ZW566 compared to YLC2-155, in agreement with its higher inhibitory activity. Mg2+ titration data also indicated a stronger dependence on Mg2+ for the RNH interaction with ZW566 compared to YLC2-155. Combined docking and quantum mechanical calculation results suggest that stronger metal coordination as well as more protein-inhibitor interactions may account for the higher binding affinity of ZW566. These findings support the idea that strategies for the development of potent competitive active site RNH inhibitors should take into account not only metal-inhibitor coordination but also protein-inhibitor interaction and conformational selectivity.
Keywords: HIV; NMR; RNase H; molecular docking; quantum mechanical calculations.
Figures
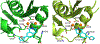




Similar articles
-
A 2-Hydroxyisoquinoline-1,3-Dione Active-Site RNase H Inhibitor Binds in Multiple Modes to HIV-1 Reverse Transcriptase.Antimicrob Agents Chemother. 2017 Sep 22;61(10):e01351-17. doi: 10.1128/AAC.01351-17. Print 2017 Oct. Antimicrob Agents Chemother. 2017. PMID: 28760905 Free PMC article.
-
Structural basis of the allosteric inhibitor interaction on the HIV-1 reverse transcriptase RNase H domain.Chem Biol Drug Des. 2012 Nov;80(5):706-16. doi: 10.1111/cbdd.12010. Epub 2012 Aug 31. Chem Biol Drug Des. 2012. PMID: 22846652 Free PMC article.
-
Entire-Dataset Analysis of NMR Fast-Exchange Titration Spectra: A Mg2+ Titration Analysis for HIV-1 Ribonuclease H Domain.J Phys Chem B. 2016 Dec 15;120(49):12420-12431. doi: 10.1021/acs.jpcb.6b08323. Epub 2016 Dec 5. J Phys Chem B. 2016. PMID: 27973819 Free PMC article.
-
Recent progress in the research of small molecule HIV-1 RNase H inhibitors.Curr Med Chem. 2014 Jun;21(17):1956-67. doi: 10.2174/0929867321666140120121158. Curr Med Chem. 2014. PMID: 24438523 Review.
-
HIV-1 integrase: the next target for AIDS therapy?Pathol Biol (Paris). 2001 Apr;49(3):237-46. doi: 10.1016/s0369-8114(01)00135-3. Pathol Biol (Paris). 2001. PMID: 11367559 Review.
Cited by
-
Strategies to overcome HIV drug resistance-current and future perspectives.Front Microbiol. 2023 Feb 16;14:1133407. doi: 10.3389/fmicb.2023.1133407. eCollection 2023. Front Microbiol. 2023. PMID: 36876064 Free PMC article. Review.
-
Avoiding Drug Resistance in HIV Reverse Transcriptase.Chem Rev. 2021 Mar 24;121(6):3271-3296. doi: 10.1021/acs.chemrev.0c00967. Epub 2021 Jan 28. Chem Rev. 2021. PMID: 33507067 Free PMC article. Review.
-
Targeting HIV-1 RNase H: N'-(2-Hydroxy-benzylidene)-3,4,5-Trihydroxybenzoylhydrazone as Selective Inhibitor Active against NNRTIs-Resistant Variants.Viruses. 2020 Jul 6;12(7):729. doi: 10.3390/v12070729. Viruses. 2020. PMID: 32640577 Free PMC article.
-
2-(Arylamino)-6-(trifluoromethyl)nicotinic Acid Derivatives: New HIV-1 RT Dual Inhibitors Active on Viral Replication.Molecules. 2020 Mar 15;25(6):1338. doi: 10.3390/molecules25061338. Molecules. 2020. PMID: 32183488 Free PMC article.
-
A Practical Approach to Bicyclic Carbamoyl Pyridones with Application to the Synthesis of HIV-1 Integrase Strand Transfer Inhibitors.Molecules. 2023 Feb 2;28(3):1428. doi: 10.3390/molecules28031428. Molecules. 2023. PMID: 36771093 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

