Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis
- PMID: 31578525
- PMCID: PMC6858560
- DOI: 10.1038/s41586-019-1618-0
Coordinated alterations in RNA splicing and epigenetic regulation drive leukaemogenesis
Abstract
Transcription and pre-mRNA splicing are key steps in the control of gene expression and mutations in genes regulating each of these processes are common in leukaemia1,2. Despite the frequent overlap of mutations affecting epigenetic regulation and splicing in leukaemia, how these processes influence one another to promote leukaemogenesis is not understood and, to our knowledge, there is no functional evidence that mutations in RNA splicing factors initiate leukaemia. Here, through analyses of transcriptomes from 982 patients with acute myeloid leukaemia, we identified frequent overlap of mutations in IDH2 and SRSF2 that together promote leukaemogenesis through coordinated effects on the epigenome and RNA splicing. Whereas mutations in either IDH2 or SRSF2 imparted distinct splicing changes, co-expression of mutant IDH2 altered the splicing effects of mutant SRSF2 and resulted in more profound splicing changes than either mutation alone. Consistent with this, co-expression of mutant IDH2 and SRSF2 resulted in lethal myelodysplasia with proliferative features in vivo and enhanced self-renewal in a manner not observed with either mutation alone. IDH2 and SRSF2 double-mutant cells exhibited aberrant splicing and reduced expression of INTS3, a member of the integrator complex3, concordant with increased stalling of RNA polymerase II (RNAPII). Aberrant INTS3 splicing contributed to leukaemogenesis in concert with mutant IDH2 and was dependent on mutant SRSF2 binding to cis elements in INTS3 mRNA and increased DNA methylation of INTS3. These data identify a pathogenic crosstalk between altered epigenetic state and splicing in a subset of leukaemias, provide functional evidence that mutations in splicing factors drive myeloid malignancy development, and identify spliceosomal changes as a mediator of IDH2-mutant leukaemogenesis.
Conflict of interest statement
Competing interests
A.M.I. has served as a consultant/advisory board member for Foundation Medicine. E.M.S. has served on advisory boards for Astellas Pharma, Daiichi Sankyo, Bayer, Novartis, Syros, Pfizer, PTC Therapeutics, AbbVie, Agios, and Celgene and has received research support from Agios, Celgene, Syros and Bayer. R.L.L. is on the Supervisory Board of Qiagen and the Scientific Advisory Board of Loxo, reports receiving commercial research grants from Celgene, Roche, and Prelude, has received honoraria from the speakers bureaus of Gilead and Lilly, has ownership interest (including stock, patents, etc.) in Qiagen and Loxo, and is a consultant/advisory board member for Novartis, Roche, Janssen, Celgene, and Incyte. A.R.K. is a founder, director, advisor, stockholder, and chair of the SAB of Stoke Therapeutics, and receives compensation from the company; A.R.K. is a paid consultant for Biogen; he is a member of the SABs of Skyhawk Therapeutics, Envisagenics BioAnalytics, and Autoimmunity Biologic Solutions, and has received compensation from these companies in the form of stock; A.R.K. is a research collaborator of Ionis Pharmaceuticals and has received royalty income from Ionis through his employer, Cold Spring Harbor Laboratory. O.A.-W. has served as a consultant for H3 Biomedicine, Foundation Medicine Inc., Merck, and Janssen; O.A.-W. has received personal speaking fees from Daiichi Sankyo. O.A.-W. has received prior research funding from H3 Biomedicine unrelated to the current manuscript. D.I., R.K.B. and O.A.-W. are inventors on a provisional patent application (patent number FHCC.P0044US.P) applied for by Fred Hutchinson Cancer Research Center on the role of reactivating BRD9 expression in cancer by modulating aberrant BRD9 splicing in
Figures












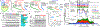

References
-
- Darman RB et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3’ Splice Site Selection through Use of a Different Branch Point. Cell Rep 13, 1033–1045 (2015). - PubMed
Online-only references
-
- Georgiades P et al. VavCre transgenic mice: a tool for mutagenesis in hematopoietic and endothelial lineages. Genesis 34, 251–256 (2002). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

