ID1 Mediates Escape from TGFβ Tumor Suppression in Pancreatic Cancer
- PMID: 31582374
- PMCID: PMC6954299
- DOI: 10.1158/2159-8290.CD-19-0529
ID1 Mediates Escape from TGFβ Tumor Suppression in Pancreatic Cancer
Abstract
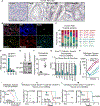
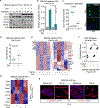
TGFβ is an important tumor suppressor in pancreatic ductal adenocarcinoma (PDA), yet inactivation of TGFβ pathway components occurs in only half of PDA cases. TGFβ cooperates with oncogenic RAS signaling to trigger epithelial-to-mesenchymal transition (EMT) in premalignant pancreatic epithelial progenitors, which is coupled to apoptosis owing to an imbalance of SOX4 and KLF5 transcription factors. We report that PDAs that develop with the TGFβ pathway intact avert this apoptotic effect via ID1. ID1 family members are expressed in PDA progenitor cells and encode components of a set of core transcriptional regulators shared by PDAs. PDA progression selects against TGFβ-mediated repression of ID1. The sustained expression of ID1 uncouples EMT from apoptosis in PDA progenitors. AKT signaling and mechanisms linked to low-frequency genetic events converge on ID1 to preserve its expression in PDA. Our results identify ID1 as a crucial node and potential therapeutic target in PDA. SIGNIFICANCE: Half of PDAs escape TGFβ-induced tumor suppression without inactivating the TGFβ pathway. We report that ID1 expression is selected for in PDAs and that ID1 uncouples TGFβ-induced EMT from apoptosis. ID1 thus emerges as a crucial regulatory node and a target of interest in PDA.This article is highlighted in the In This Issue feature, p. 1.
©2019 American Association for Cancer Research.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

