Centrosomal and ciliary targeting of CCDC66 requires cooperative action of centriolar satellites, microtubules and molecular motors
- PMID: 31582766
- PMCID: PMC6776500
- DOI: 10.1038/s41598-019-50530-4
Centrosomal and ciliary targeting of CCDC66 requires cooperative action of centriolar satellites, microtubules and molecular motors
Abstract
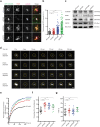
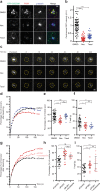
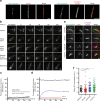
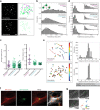
Mammalian centrosomes and cilia play key roles in many cellular processes and their deregulation is linked to cancer and ciliopathies. Spatiotemporal regulation of their biogenesis and function in response to physiological stimuli requires timely protein targeting. This can occur by different pathways, including microtubule-dependent active transport and via centriolar satellites, which are key regulators of cilia assembly and signaling. How satellites mediate their functions and their relationship with other targeting pathways is currently unclear. To address this, we studied retinal degeneration gene product CCDC66, which localizes to centrosomes, cilia, satellites and microtubules and functions in ciliogenesis. FRAP experiments showed that its centrosomal pool was dynamic and the ciliary pool associated with the ciliary axoneme and was stable. Centrosomal CCDC66 abundance and dynamics required microtubule-dependent active transport and tethering, and was inhibited by sequestration at satellites. Systematic quantitation of satellite dynamics identified only a small fraction to display microtubule-based bimodal motility, consistent with trafficking function. Majority displayed diffusive motility with unimodal persistence, supporting sequestration function. Together, our findings reveal new mechanisms of communication between membrane-less compartments.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

