Dynamics and genomic landscape of CD8+ T cells undergoing hepatic priming
- PMID: 31582858
- PMCID: PMC6858885
- DOI: 10.1038/s41586-019-1620-6
Dynamics and genomic landscape of CD8+ T cells undergoing hepatic priming
Abstract
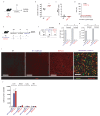
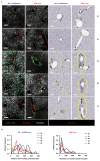
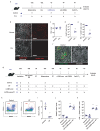
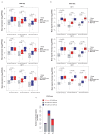
The responses of CD8+ T cells to hepatotropic viruses such as hepatitis B range from dysfunction to differentiation into effector cells, but the mechanisms that underlie these distinct outcomes remain poorly understood. Here we show that priming by Kupffer cells, which are not natural targets of hepatitis B, leads to differentiation of CD8+ T cells into effector cells that form dense, extravascular clusters of immotile cells scattered throughout the liver. By contrast, priming by hepatocytes, which are natural targets of hepatitis B, leads to local activation and proliferation of CD8+ T cells but not to differentiation into effector cells; these cells form loose, intravascular clusters of motile cells that coalesce around portal tracts. Transcriptomic and chromatin accessibility analyses reveal unique features of these dysfunctional CD8+ T cells, with limited overlap with those of exhausted or tolerant T cells; accordingly, CD8+ T cells primed by hepatocytes cannot be rescued by treatment with anti-PD-L1, but instead respond to IL-2. These findings suggest immunotherapeutic strategies against chronic hepatitis B infection.
Conflict of interest statement
The authors declare competing financial interest: M.I., L.G.G., R.O., A.C. and L.N. are inventors on patents filed, owned and managed by Telethon Foundation and San Raffaele Scientific Institute on LV technology related to the work presented in this manuscript. Readers are welcome to comment on the online version of the paper.
Figures



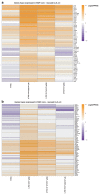


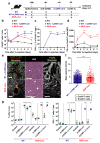
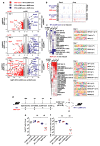

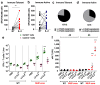
References
-
- Warren A, et al. T lymphocytes interact with hepatocytes through fenestrations in murine liver sinusoidal endothelial cells. Hepatology. 2006;44:1182–1190. - PubMed
-
- Wong YC, Tay SS, McCaughan GW, Bowen DG, Bertolino P. Immune outcomes in the liver: Is CD8 T cell fate determined by the environment? J Hepatol. 2015;63:1005–1014. - PubMed
-
- Holz LE, et al. Naïve CD8 T cell activation by liver bone marrow-derived cells leads to a ‘neglected’ IL-2low Bimhigh phenotype, poor CTL function and cell death. J Hepatol. 2012;57:830–836. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

