DNA Methylation Validation Methods: a Coherent Review with Practical Comparison
- PMID: 31582911
- PMCID: PMC6771119
- DOI: 10.1186/s12575-019-0107-z
DNA Methylation Validation Methods: a Coherent Review with Practical Comparison
Abstract
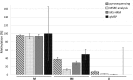
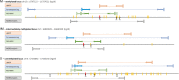
Here, we present a practical overview of four commonly used validation methods for DNA methylation assessment: methylation specific restriction endonucleases (MSRE) analysis, pyrosequencing, methylation specific high-resolution DNA melting (MS-HRM) and quantitative methylation specific polymerase chain reaction (qMSP). Using these methods, we measured DNA methylation levels of three loci in human genome among which one was highly methylated, one intermediately methylated and one unmethylated. We compared the methods in terms of primer design demands, methods' feasibility, accuracy, time and money consumption, and usability for clinical diagnostics. Pyrosequencing and MS-HRM proved to be the most convenient methods. Using pyrosequencing, it is possible to analyze every CpG in a chosen region. The price of the instrument may represent the main limitation of this methodology. MS-HRM is a simple PCR-based method. The measurement was quick, cheap and very accurate. MSRE analysis is based on a methylation specific digestion of DNA. It does not require a bisulfite conversion of DNA as the other methods. MSRE analysis was very easy to perform, however, it was not suitable for intermediately methylated regions and it was also quite expensive. qMSP is a qPCR-based method that uses primers designed specifically for methylated and unmethylated alleles of a chosen region. This was the least accurate method and also the primer design and optimization of PCR conditions were highly demanding.
Keywords: DNA methylation; MS-HRM; MSRE; Pyrosequencing; Validation methods; qMSP.
© The Author(s). 2019.
Conflict of interest statement
Competing InterestsThe authors declare that they have no competing interests.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

