PDBe-KB: a community-driven resource for structural and functional annotations
- PMID: 31584092
- PMCID: PMC6943075
- DOI: 10.1093/nar/gkz853
PDBe-KB: a community-driven resource for structural and functional annotations
Abstract
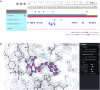
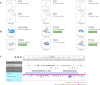
The Protein Data Bank in Europe-Knowledge Base (PDBe-KB, https://pdbe-kb.org) is a community-driven, collaborative resource for literature-derived, manually curated and computationally predicted structural and functional annotations of macromolecular structure data, contained in the Protein Data Bank (PDB). The goal of PDBe-KB is two-fold: (i) to increase the visibility and reduce the fragmentation of annotations contributed by specialist data resources, and to make these data more findable, accessible, interoperable and reusable (FAIR) and (ii) to place macromolecular structure data in their biological context, thus facilitating their use by the broader scientific community in fundamental and applied research. Here, we describe the guidelines of this collaborative effort, the current status of contributed data, and the PDBe-KB infrastructure, which includes the data exchange format, the deposition system for added value annotations, the distributable database containing the assembled data, and programmatic access endpoints. We also describe a series of novel web-pages-the PDBe-KB aggregated views of structure data-which combine information on macromolecular structures from many PDB entries. We have recently released the first set of pages in this series, which provide an overview of available structural and functional information for a protein of interest, referenced by a UniProtKB accession.
© The Author(s) 2019. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
- U01 CA239106/CA/NCI NIH HHS/United States
- BB/P023959/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R01 GM114409/GM/NIGMS NIH HHS/United States
- BB/H018409/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 23187/CRUK_/Cancer Research UK/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- BB/G022682/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/L01257X/1/MRC_/Medical Research Council/United Kingdom
- BBS/B/16542/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/L020742/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- NH/12/2/29427/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources

