Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato
- PMID: 31584235
- PMCID: PMC7061876
- DOI: 10.1111/pbi.13272
Crosstalk of PIF4 and DELLA modulates CBF transcript and hormone homeostasis in cold response in tomato
Abstract
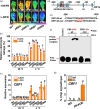
The ability to interpret daily and seasonal fluctuations, latitudinal and vegetation canopy variations in light and temperature signals is essential for plant survival. However, the precise molecular mechanisms transducing the signals from light and temperature perception to maintain plant growth and adaptation remain elusive. We show that far-red light induces PHYTOCHROME-INTERACTING TRANSCRIPTION 4 (SlPIF4) accumulation under low-temperature conditions via phytochrome A in Solanum lycopersicum (tomato). Reverse genetic approaches revealed that knocking out SlPIF4 increases cold susceptibility, while overexpressing SlPIF4 enhances cold tolerance in tomato plants. SlPIF4 not only directly binds to the promoters of the C-REPEAT BINDING FACTOR (SlCBF) genes and activates their expression but also regulates plant hormone biosynthesis and signals, including abscisic acid, jasmonate and gibberellin (GA), in response to low temperature. Moreover, SlPIF4 directly activates the SlDELLA gene (GA-INSENSITIVE 4, SlGAI4) under cold stress, and SlGAI4 positively regulates cold tolerance. Additionally, SlGAI4 represses accumulation of the SlPIF4 protein, thus forming multiple coherent feed-forward loops. Our results reveal that plants integrate light and temperature signals to better adapt to cold stress through shared hormone pathways and transcriptional regulators, which may provide a comprehensive understanding of plant growth and survival in a changing environment.
Keywords: GAI4; PIF4; Solanum lycopersicum (tomato); cold stress; hormone; light signalling.
© 2019 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interests.
Figures









References
-
- Achard, P. , Cheng, H. , De Grauwe, L. , Decat, J. , Schoutteten, H. , Moritz, T. , Van Der Straeten, D. et al. (2006) Integration of plant responses to environmentally activated phytohormonal signals. Science, 311, 91–94. - PubMed
-
- Bradford, M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal. Biochem. 72, 248–254. - PubMed
-
- Casal, J.J. and Qüesta, J.I. (2018) Light and temperature cues: multitasking receptors and transcriptional integrators. New Phytol. 217, 1029–1034. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

