2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records
- PMID: 31584655
- PMCID: PMC7489085
- DOI: 10.1093/jamia/ocz166
2018 n2c2 shared task on adverse drug events and medication extraction in electronic health records
Abstract
Objective: This article summarizes the preparation, organization, evaluation, and results of Track 2 of the 2018 National NLP Clinical Challenges shared task. Track 2 focused on extraction of adverse drug events (ADEs) from clinical records and evaluated 3 tasks: concept extraction, relation classification, and end-to-end systems. We perform an analysis of the results to identify the state of the art in these tasks, learn from it, and build on it.
Materials and methods: For all tasks, teams were given raw text of narrative discharge summaries, and in all the tasks, participants proposed deep learning-based methods with hand-designed features. In the concept extraction task, participants used sequence labelling models (bidirectional long short-term memory being the most popular), whereas in the relation classification task, they also experimented with instance-based classifiers (namely support vector machines and rules). Ensemble methods were also popular.
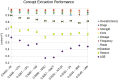
Results: A total of 28 teams participated in task 1, with 21 teams in tasks 2 and 3. The best performing systems set a high performance bar with F1 scores of 0.9418 for concept extraction, 0.9630 for relation classification, and 0.8905 for end-to-end. However, the results were much lower for concepts and relations of Reasons and ADEs. These were often missed because local context is insufficient to identify them.
Conclusions: This challenge shows that clinical concept extraction and relation classification systems have a high performance for many concept types, but significant improvement is still required for ADEs and Reasons. Incorporating the larger context or outside knowledge will likely improve the performance of future systems.
© The Author(s) 2019. Published by Oxford University Press on behalf of the American Medical Informatics Association. All rights reserved. For permissions, please email: journals.permissions@oup.com.
Figures
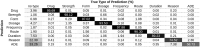


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

