The SAUR41 subfamily of SMALL AUXIN UP RNA genes is abscisic acid inducible to modulate cell expansion and salt tolerance in Arabidopsis thaliana seedlings
- PMID: 31585004
- PMCID: PMC7182593
- DOI: 10.1093/aob/mcz160
The SAUR41 subfamily of SMALL AUXIN UP RNA genes is abscisic acid inducible to modulate cell expansion and salt tolerance in Arabidopsis thaliana seedlings
Abstract
Background and aims: Most primary auxin response genes are classified into three families: AUX/IAA, GH3 and SAUR genes. Few studies have been conducted on Arabidopsis thaliana SAUR genes, possibly due to genetic redundancy among different subfamily members. Data mining on arabidopsis transcriptional profiles indicates that the SAUR41 subfamily members of SMALL AUXIN UP RNA genes are, strikingly, induced by an inhibitory phytohormone, abscisic acid (ABA). We aimed to reveal the physiological roles of arabidopsis SAUR41 subfamily genes containing SAUR40, SAUR41, SAUR71 and SAUR72.
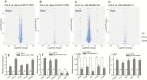
Methods: Transcriptional responses of arabidopsis SAUR41 genes to phytohormones were determined by quantitative real-time PCR. Knock out of SAUR41 genes was carried out with the CRISPR/Cas9 (clustered regulatory interspaced short palindromic repeats/CRISPR-associated protein 9) genome editing technique. The saur41/40/71/72 quadruple mutants, SAUR41 overexpression lines and the wild type were subjected to ultrastructural observation, transcriptome analysis and physiological characterization.
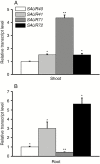
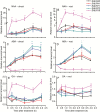
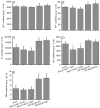
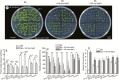
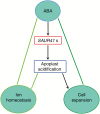
Key results: Transcription of arabidopsis SAUR41 subfamily genes is activated by ABA but not by gibberellic acids and brassinosteroids. Quadruple mutations in saur41/40/71/72 led to reduced cell expansion/elongation in cotyledons and hypocotyls, opposite to the overexpression of SAUR41; however, an irregular arrangement of cell size and shape was observed in both cases. The quadruple mutants had increased transcription of calcium homeostasis/signalling genes in seedling shoots, and the SAUR41 overexpression lines had decreased transcription of iron homeostasis genes in roots and increased ABA biosynthesis in shoots. Notably, both the quadruple mutants and the SAUR41 overexpression lines were hypersensitive to salt stress during seedling establishment, whereas specific expression of SAUR41 under the ABA-responsive RD29A (Responsive to Desiccation 29A) promoter in the quadruple mutants rescued the inhibitory effect of salt stress.
Conclusions: The SAUR41 subfamily genes of arabidopsis are ABA inducible to modulate cell expansion, ion homeostasis and salt tolerance. Our work may provide new candidate genes for improvement of plant abiotic stress tolerance.
Keywords: Arabidopsis thaliana; SMALL AUXIN UP RNA genes; CRISPR/Cas9; abscisic acid; cell expansion; ion homeostasis; salt tolerance; transcription profiling.
© The Author(s) 2019. Published by Oxford University Press on behalf of the Annals of Botany Company. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures




Similar articles
-
The SAUR41 subfamily of cell expansion-promoting genes modulates abscisic acid sensitivity and root touch response: a possible connection to ion homeostasis regulation.Plant Signal Behav. 2020;15(1):1702239. doi: 10.1080/15592324.2019.1702239. Epub 2019 Dec 10. Plant Signal Behav. 2020. PMID: 31822155 Free PMC article.
-
The tissue-specific and developmentally regulated expression patterns of the SAUR41 subfamily of small auxin up RNA genes: potential implications.Plant Signal Behav. 2013 Aug;8(8):e25283. doi: 10.4161/psb.25283. Epub 2013 Jun 10. Plant Signal Behav. 2013. PMID: 23759547 Free PMC article.
-
Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis.Plant Cell Physiol. 2013 Apr;54(4):609-21. doi: 10.1093/pcp/pct028. Epub 2013 Feb 8. Plant Cell Physiol. 2013. PMID: 23396598
-
Overexpression of HbMBF1a, encoding multiprotein bridging factor 1 from the halophyte Hordeum brevisubulatum, confers salinity tolerance and ABA insensitivity to transgenic Arabidopsis thaliana.Plant Mol Biol. 2020 Jan;102(1-2):1-17. doi: 10.1007/s11103-019-00926-7. Epub 2019 Oct 26. Plant Mol Biol. 2020. PMID: 31655970 Free PMC article.
-
The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants.Planta. 2017 Dec;246(6):1215-1231. doi: 10.1007/s00425-017-2766-9. Epub 2017 Aug 31. Planta. 2017. PMID: 28861611
Cited by
-
Gene Editing for Plant Resistance to Abiotic Factors: A Systematic Review.Plants (Basel). 2023 Jan 9;12(2):305. doi: 10.3390/plants12020305. Plants (Basel). 2023. PMID: 36679018 Free PMC article. Review.
-
Engineering drought and salinity tolerance traits in crops through CRISPR-mediated genome editing: Targets, tools, challenges, and perspectives.Plant Commun. 2022 Nov 14;3(6):100417. doi: 10.1016/j.xplc.2022.100417. Epub 2022 Aug 3. Plant Commun. 2022. PMID: 35927945 Free PMC article. Review.
-
The Arabidopsis SMALL AUXIN UP RNA32 Protein Regulates ABA-Mediated Responses to Drought Stress.Front Plant Sci. 2021 Mar 12;12:625493. doi: 10.3389/fpls.2021.625493. eCollection 2021. Front Plant Sci. 2021. PMID: 33777065 Free PMC article.
-
The SAUR41 subfamily of cell expansion-promoting genes modulates abscisic acid sensitivity and root touch response: a possible connection to ion homeostasis regulation.Plant Signal Behav. 2020;15(1):1702239. doi: 10.1080/15592324.2019.1702239. Epub 2019 Dec 10. Plant Signal Behav. 2020. PMID: 31822155 Free PMC article.
-
Involvement of Auxin-Mediated CqEXPA50 Contributes to Salt Tolerance in Quinoa (Chenopodium quinoa) by Interaction with Auxin Pathway Genes.Int J Mol Sci. 2022 Jul 30;23(15):8480. doi: 10.3390/ijms23158480. Int J Mol Sci. 2022. PMID: 35955612 Free PMC article.
References
-
- Arsuffi G, Braybrook SA. 2017. Acid growth: an ongoing trip. Journal of Experimental Botany 69: 137–146. - PubMed
-
- Bechtold N. 1993. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. Compte Rendu de l’Academie des Sciences Paris, Sciences de la Vie 316: 1194–1199.
-
- Chae K, Isaacs CG, Reeves PH, et al. . 2012. Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. The Plant Journal 71: 684–697. - PubMed
-
- Cutler SR, Rodriguez PL, Finkelstein RR. 2010. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology 61: 651–679. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

