Diagnostic Utility of Next-Generation Sequencing for Disorders of Somatic Mosaicism: A Five-Year Cumulative Cohort
- PMID: 31585106
- PMCID: PMC6817554
- DOI: 10.1016/j.ajhg.2019.09.002
Diagnostic Utility of Next-Generation Sequencing for Disorders of Somatic Mosaicism: A Five-Year Cumulative Cohort
Abstract
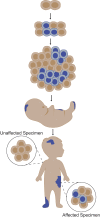
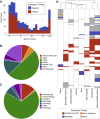
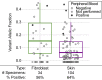
Disorders of somatic mosaicism (DoSM) are a diverse group of syndromic and non-syndromic conditions caused by mosaic variants in genes that regulate cell survival and proliferation. Despite overlap in gene space and technical requirements, few clinical labs specialize in DoSM compared to oncology. We adapted a high-sensitivity next-generation sequencing cancer assay for DoSM in 2014. Some 343 individuals have been tested over the past 5 years, 58% of which had pathogenic and likely pathogenic (P/LP) findings, for a total of 206 P/LP variants in 22 genes. Parameters associated with the high diagnostic yield were: (1) deep sequencing (∼2,000× coverage), (2) a broad gene set, and (3) testing affected tissues. Fresh and formalin-fixed paraffin embedded tissues performed equivalently for identification of P/LP variants (62% and 71% of individuals, respectively). Comparing cultured fibroblasts to skin biopsies suggested that culturing might boost the allelic fraction of variants that confer a growth advantage, specifically gain-of-function variants in PIK3CA. Buccal swabs showed high diagnostic sensitivity in case subjects where disease phenotypes manifested in the head or brain. Peripheral blood was useful as an unaffected comparator tissue to determine somatic versus constitutional origin but had poor diagnostic sensitivity. Descriptions of all tested individuals, specimens, and P/LP variants included in this cohort are available to further the study of the DoSM population.
Keywords: brain malformation; macrocephaly; megalencephaly; mosaicism; nevus syndrome; overgrowth syndrome; somatic overgrowth; vascular malformation.
Copyright © 2019. Published by Elsevier Inc.
Conflict of interest statement
B.A.D. serves as a consultant and member of the medical advisory board for Venthera. M.C.S. and J.W.H. consult for Pierian Diagnostics.
Figures



References
-
- Kinsler V.A., Thomas A.C., Ishida M., Bulstrode N.W., Loughlin S., Hing S., Chalker J., McKenzie K., Abu-Amero S., Slater O. Multiple congenital melanocytic nevi and neurocutaneous melanosis are caused by postzygotic mutations in codon 61 of NRAS. J. Invest. Dermatol. 2013;133:2229–2236. - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

