Identification of a tumor-specific allo-HLA-restricted γδTCR
- PMID: 31585951
- PMCID: PMC6784524
- DOI: 10.1182/bloodadvances.2019032409
Identification of a tumor-specific allo-HLA-restricted γδTCR
Abstract
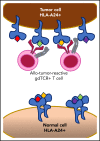
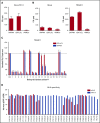
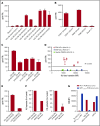
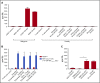
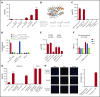
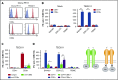
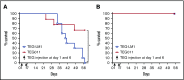
γδT cells are key players in cancer immune surveillance because of their ability to recognize malignant transformed cells, which makes them promising therapeutic tools in the treatment of cancer. However, the biological mechanisms of how γδT-cell receptors (TCRs) interact with their ligands are poorly understood. Within this context, we describe the novel allo-HLA-restricted and CD8α-dependent Vγ5Vδ1TCR. In contrast to the previous assumption of the general allo-HLA reactivity of a minor fraction of γδTCRs, we show that classic anti-HLA-directed, γδTCR-mediated reactivity can selectively act on hematological and solid tumor cells, while not harming healthy tissues in vitro and in vivo. We identified the molecular interface with proximity to the peptide-binding groove of HLA-A*24:02 as the essential determinant for recognition and describe the critical role of CD8 as a coreceptor. We conclude that alloreactive γδT-cell repertoires provide therapeutic opportunities, either within the context of haplotransplantation or as individual γδTCRs for genetic engineering of tumor-reactive T cells.
© 2019 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: G.J.J.K., D.X.B., Z.S., and J.K. are inventors on different patents with γδTCR sequences, recognition mechanisms, and isolation strategies. G.J.J.K. is employed by Kiadis Pharma. J.K. is scientific advisor and shareholder of Gadeta. The remaining authors declare no competing financial interests.
Figures

References
-
- Girardi M, Oppenheim DE, Steele CR, et al. . Regulation of cutaneous malignancy by gammadelta T cells. Science. 2001;294(5542):605-609. - PubMed
-
- Scheper W, van Dorp S, Kersting S, et al. . γδT cells elicited by CMV reactivation after allo-SCT cross-recognize CMV and leukemia. Leukemia. 2013;27(6):1328-1338. - PubMed
-
- Lalor SJ, McLoughlin RM. Memory γδ T Cells-Newly Appreciated Protagonists in Infection and Immunity. Trends Immunol. 2016;37(10):690-702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

