Mechanically activated Piezo1 channels of cardiac fibroblasts stimulate p38 mitogen-activated protein kinase activity and interleukin-6 secretion
- PMID: 31586031
- PMCID: PMC6873183
- DOI: 10.1074/jbc.RA119.009167
Mechanically activated Piezo1 channels of cardiac fibroblasts stimulate p38 mitogen-activated protein kinase activity and interleukin-6 secretion
Abstract
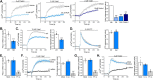
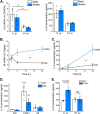
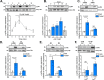
Piezo1 is a mechanosensitive cation channel with widespread physiological importance; however, its role in the heart is poorly understood. Cardiac fibroblasts help preserve myocardial integrity and play a key role in regulating its repair and remodeling following stress or injury. Here we investigated Piezo1 expression and function in cultured human and mouse cardiac fibroblasts. RT-PCR experiments confirmed that Piezo1 mRNA in cardiac fibroblasts is expressed at levels similar to those in endothelial cells. The results of a Fura-2 intracellular Ca2+ assay validated Piezo1 as a functional ion channel that is activated by its agonist, Yoda1. Yoda1-induced Ca2+ entry was inhibited by Piezo1 blockers (gadolinium and ruthenium red) and was reduced proportionally by siRNA-mediated Piezo1 knockdown or in murine Piezo1+/- cells. Results from cell-attached patch clamp recordings on human cardiac fibroblasts established that they contain mechanically activated ion channels and that their pressure responses are reduced by Piezo1 knockdown. Investigation of Yoda1 effects on selected remodeling genes indicated that Piezo1 activation increases both mRNA levels and protein secretion of IL-6, a pro-hypertrophic and profibrotic cytokine, in a Piezo1-dependent manner. Moreover, Piezo1 knockdown reduced basal IL-6 expression from cells cultured on softer collagen-coated substrates. Multiplex kinase activity profiling combined with kinase inhibitor experiments and phosphospecific immunoblotting established that Piezo1 activation stimulates IL-6 secretion via the p38 mitogen-activated protein kinase downstream of Ca2+ entry. In summary, cardiac fibroblasts express mechanically activated Piezo1 channels coupled to secretion of the paracrine signaling molecule IL-6. Piezo1 may therefore be important in regulating cardiac remodeling.
Keywords: IL-6; calcium; cardiac fibroblast; fibroblast; heart; ion channel; mechanotransduction; mitogen-activated protein kinase (MAPK); p38 MAPK; patch clamp; signal transduction.
© 2019 Blythe et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

