Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance
- PMID: 31586049
- PMCID: PMC6778101
- DOI: 10.1038/s41467-019-12364-6
Integrated evolutionary analysis reveals antimicrobial peptides with limited resistance
Abstract
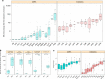
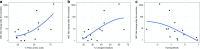
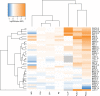
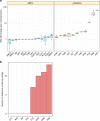
Antimicrobial peptides (AMPs) are promising antimicrobials, however, the potential of bacterial resistance is a major concern. Here we systematically study the evolution of resistance to 14 chemically diverse AMPs and 12 antibiotics in Escherichia coli. Our work indicates that evolution of resistance against certain AMPs, such as tachyplesin II and cecropin P1, is limited. Resistance level provided by point mutations and gene amplification is very low and antibiotic-resistant bacteria display no cross-resistance to these AMPs. Moreover, genomic fragments derived from a wide range of soil bacteria confer no detectable resistance against these AMPs when introduced into native host bacteria on plasmids. We have found that simple physicochemical features dictate bacterial propensity to evolve resistance against AMPs. Our work could serve as a promising source for the development of new AMP-based therapeutics less prone to resistance, a feature necessary to avoid any possible interference with our innate immune system.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Yeaman, M. R. & Yount, N. Y. Mechanisms of antimicrobial peptide action and resistance. 57, 1–24 (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

